Reshaping data with tidyr
Annoucements
- Midterm II is Thursday, November 14th.
- Material on the midterm will include all material through Week 11.
Agenda
- Continuation and review of combining tibbles (i.e. data joins)
- Reshaping data with
tidyr
Data Joins
Data Joins in R
Recall the x and y tibbles from last time.
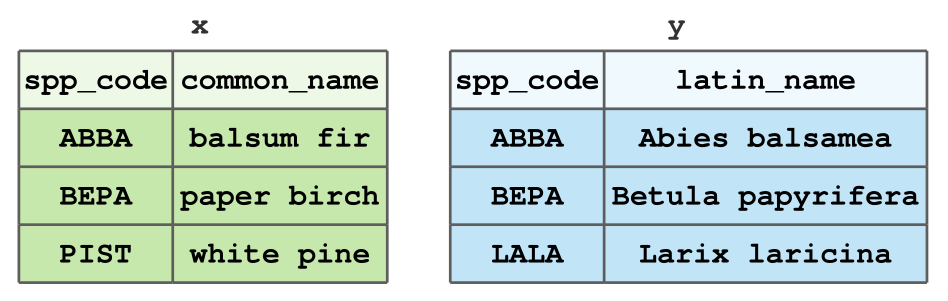
Types of Data Joins
The dplyr package includes functions for two general types of joins:
- Mutating joins, which combine the columns of tibbles
xandy, and - Filtering joins, which match the rows of tibbles
xandy.
Think of how mutate() adds columns to a tibble, while filter() removes rows.
Mutating Joins
dplyr contains four mutating joins:
left_join(x, y)keeps all rows ofx, but if a row inydoes not match tox, anNAis assigned to that row in the new columns.right_join(x, y)is equivalent toleft_join(y, x), except for column order.inner_join(x, y)keeps only the rows matched betweenxandy.full_join(x, y)keeps all rows of bothxandy.
Filtering Joins
dplyr contains two filtering joins:
semi_join(x, y)keeps all the rows inxthat have a match iny.anti_join(x, y)removes all the rows inxthat have a match iny.
Note: Unlike mutating joins, filtering joins do not add any columns to the data.
Loading data
We’ll create the x and y tibbles in R:
Loading data
We’ll create the x and y tibbles in R:
Q1. Write the code to get this output
# A tibble: 3 × 3
spp_code common_name latin_name
<chr> <chr> <chr>
1 ABBA balsum fir Abies balsamea
2 BEPA paper birch Betula papyrifera
3 PIST white pine <NA> 01:00
Q1. Write the code to get this output
Q2. Write the code to get this output
# A tibble: 4 × 3
spp_code common_name latin_name
<chr> <chr> <chr>
1 ABBA balsum fir Abies balsamea
2 BEPA paper birch Betula papyrifera
3 PIST white pine <NA>
4 LALA <NA> Larix laricina 01:00
Q2. Write the code to get this output
Q3. Write the code to get this output
# A tibble: 3 × 3
spp_code latin_name common_name
<chr> <chr> <chr>
1 ABBA Abies balsamea balsum fir
2 BEPA Betula papyrifera paper birch
3 LALA Larix laricina <NA> 01:00
Q3. Write the code to get this output
Q4. Write the code to get this output
# A tibble: 3 × 3
spp_code common_name latin_name
<chr> <chr> <chr>
1 ABBA balsum fir Abies balsamea
2 BEPA paper birch Betula papyrifera
3 LALA <NA> Larix laricina 01:00
Q4. Write the code to get this output
Q5. Write the code to get this output
# A tibble: 2 × 3
spp_code common_name latin_name
<chr> <chr> <chr>
1 ABBA balsum fir Abies balsamea
2 BEPA paper birch Betula papyrifera01:00
Q5. Write the code to get this output
Q6. Write the code to get this output
# A tibble: 4 × 3
spp_code latin_name common_name
<chr> <chr> <chr>
1 ABBA Abies balsamea balsum fir
2 BEPA Betula papyrifera paper birch
3 LALA Larix laricina <NA>
4 PIST <NA> white pine 01:00
Q6. Write the code to get this output
Consider a third tibble, z:
Q7. Write the code to get this output
# A tibble: 3 × 3
species_code common_name latin_name
<chr> <chr> <chr>
1 ABBA balsum fir Abies balsamea
2 BEPA paper birch Betula papyrifera
3 PIST white pine <NA> 01:00
Q7. Write the code to get this output
Q8. Write the code to get this output (hint: use a filtering join)
# A tibble: 2 × 2
spp_code latin_name
<chr> <chr>
1 ABBA Abies balsamea
2 BEPA Betula papyrifera01:00
Q8. Write the code to get this output (hint: use a filtering join)
Q9. Write the code to get this output (hint: use a filtering join)
# A tibble: 1 × 2
species_code common_name
<chr> <chr>
1 PIST white pine 01:00
Q9. Write the code to get this output (hint: use a filtering join)
Q10. Write the code to get this output (hint: use a filtering join)
# A tibble: 1 × 2
spp_code latin_name
<chr> <chr>
1 LALA Larix laricina01:00
Q10. Write the code to get this output (hint: use a filtering join)
Reshaping Data
The tidyr package
- The
tidyrpackage allows us to transform data from long to wide formats, and back. - A key use of the
tidyrpackage is getting data in tidy format. - But what is tidy format?
Tidy data
“Tidy data” or “tidy format” is a formal concept of how we organize data for analyses, in particular, with tidy data:
- Each row of the data correspond to a single observation, and
- Each column of the data correspond to a variable.
Tidy data
“Tidy data” or “tidy format” is a formal concept of how we organize data for analyses, in particular, with tidy data:
- Each row of the data correspond to a single observation, and
- Each column of the data correspond to a variable.
Let’s take a look at a blog post from Julie Lowndes and Allison Horst about tidy data
Tidying data with tidyr
Consider the face dataset from IFDAR:
face <- read_csv("../labs/datasets/FACE/FACE_aspen_core_growth.csv") %>%
select(Rep, Treat, Clone, ID = `ID #`,
contains(as.character(2001:2005)) & contains("Height"))
face# A tibble: 1,991 × 9
Rep Treat Clone ID `2001_Height` `2002_Height` `2003_Height`
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 8 45 NA NA NA
2 1 1 216 44 547 622 715
3 1 1 8 43 273 275 305
4 1 1 216 42 526 619 720
5 1 1 216 54 328 341 364
6 1 1 271 55 543 590 634
7 1 1 271 56 450 502 587
8 1 1 8 57 217 227 256
9 1 1 259 58 158 155 NA
10 1 1 271 59 230 241 260
# ℹ 1,981 more rows
# ℹ 2 more variables: `2004_Height` <dbl>, `2005_Height` <dbl>Tidying data with tidyr
Is this data tidy? If so, what does a row represent? What does a column represent?
# A tibble: 1,991 × 9
Rep Treat Clone ID `2001_Height` `2002_Height` `2003_Height`
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 8 45 NA NA NA
2 1 1 216 44 547 622 715
3 1 1 8 43 273 275 305
4 1 1 216 42 526 619 720
5 1 1 216 54 328 341 364
6 1 1 271 55 543 590 634
7 1 1 271 56 450 502 587
8 1 1 8 57 217 227 256
9 1 1 259 58 158 155 NA
10 1 1 271 59 230 241 260
# ℹ 1,981 more rows
# ℹ 2 more variables: `2004_Height` <dbl>, `2005_Height` <dbl>How do we make this data tidy? 🤔
# A tibble: 1,991 × 9
Rep Treat Clone ID `2001_Height` `2002_Height` `2003_Height`
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 8 45 NA NA NA
2 1 1 216 44 547 622 715
3 1 1 8 43 273 275 305
4 1 1 216 42 526 619 720
5 1 1 216 54 328 341 364
6 1 1 271 55 543 590 634
7 1 1 271 56 450 502 587
8 1 1 8 57 217 227 256
9 1 1 259 58 158 155 NA
10 1 1 271 59 230 241 260
# ℹ 1,981 more rows
# ℹ 2 more variables: `2004_Height` <dbl>, `2005_Height` <dbl>Each row should represent a measurement for a given rep, treatment, clone, and ID. But currently, we have multiple measurements on the same row.
Further, we have multiple columns for the “height” variable.
Not tidy!
We need to pivot the data into a longer format!
Enter, pivot_longer().
pivot_longer(), a function from tidyr takes four key arguments:
.data: the data (tibble) you’d like to pivot,cols: the columns you’d like to pivot,names_to: the new column that will be created which takes the column names fromcolsas values, andvalues_to: the new column that will be created which takes the column values fromcolsas values.
Why the long face?
Why the long face?
Why the long face?
- A cleaner way to select these columns is to use
dplyr’scontains()function.
Why the long face?
# A tibble: 9,955 × 6
Rep Treat Clone ID Year_Type Height_cm
<dbl> <dbl> <dbl> <dbl> <chr> <dbl>
1 1 1 8 45 2001_Height NA
2 1 1 8 45 2002_Height NA
3 1 1 8 45 2003_Height NA
4 1 1 8 45 2004_Height NA
5 1 1 8 45 2005_Height NA
6 1 1 216 44 2001_Height 547
7 1 1 216 44 2002_Height 622
8 1 1 216 44 2003_Height 715
9 1 1 216 44 2004_Height 716
10 1 1 216 44 2005_Height 817
# ℹ 9,945 more rows- To get tidy data!
Going (back) to wide data
- Sometimes, we need to “widen” a dataset to get it into tidy format.
- For this example, we will just widen the
face_longdataset back to its original form. tidyrhas an aptly named function,pivot_wider().- Key arguments of
pivot_wider():.data: the tibble to widen,names_from: the column that contains values which will be assigned as the new column names,values_from: the column that contains
Pivoting wider
face_wide <- face_long %>%
pivot_wider(names_from = "Year_Type", values_from = "Height_cm")
all_equal(face, face_wide)[1] TRUE- This results in the same tibble that we started with!
Other useful tidyr functions
unite()for pasteing column values together with specified seperators,- The
separate_wider_*()family: for splitting columns into multiple new columns:separate_wider_delim(): separate by delimiterseparate_wider_position(): separate by positionseparate_wider_regex(): separate by regular expression
unite(): examples
# A tibble: 9,955 × 6
Rep Treat Clone ID Year_Type Height_cm
<dbl> <dbl> <dbl> <dbl> <chr> <dbl>
1 1 1 8 45 2001_Height NA
2 1 1 8 45 2002_Height NA
3 1 1 8 45 2003_Height NA
4 1 1 8 45 2004_Height NA
5 1 1 8 45 2005_Height NA
6 1 1 216 44 2001_Height 547
7 1 1 216 44 2002_Height 622
8 1 1 216 44 2003_Height 715
9 1 1 216 44 2004_Height 716
10 1 1 216 44 2005_Height 817
# ℹ 9,945 more rowsunite(): examples
# A tibble: 9,955 × 4
Design ID Year_Type Height_cm
<chr> <dbl> <chr> <dbl>
1 1_1_8 45 2001_Height NA
2 1_1_8 45 2002_Height NA
3 1_1_8 45 2003_Height NA
4 1_1_8 45 2004_Height NA
5 1_1_8 45 2005_Height NA
6 1_1_216 44 2001_Height 547
7 1_1_216 44 2002_Height 622
8 1_1_216 44 2003_Height 715
9 1_1_216 44 2004_Height 716
10 1_1_216 44 2005_Height 817
# ℹ 9,945 more rowsunite(): examples
# A tibble: 9,955 × 4
Design ID Year_Type Height_cm
<chr> <dbl> <chr> <dbl>
1 1.1.8 45 2001_Height NA
2 1.1.8 45 2002_Height NA
3 1.1.8 45 2003_Height NA
4 1.1.8 45 2004_Height NA
5 1.1.8 45 2005_Height NA
6 1.1.216 44 2001_Height 547
7 1.1.216 44 2002_Height 622
8 1.1.216 44 2003_Height 715
9 1.1.216 44 2004_Height 716
10 1.1.216 44 2005_Height 817
# ℹ 9,945 more rowsunite(): examples
# A tibble: 9,955 × 4
Design ID Year_Type Height_cm
<chr> <dbl> <chr> <dbl>
1 1abc1abc8 45 2001_Height NA
2 1abc1abc8 45 2002_Height NA
3 1abc1abc8 45 2003_Height NA
4 1abc1abc8 45 2004_Height NA
5 1abc1abc8 45 2005_Height NA
6 1abc1abc216 44 2001_Height 547
7 1abc1abc216 44 2002_Height 622
8 1abc1abc216 44 2003_Height 715
9 1abc1abc216 44 2004_Height 716
10 1abc1abc216 44 2005_Height 817
# ℹ 9,945 more rowsseparate_wider_delim() examples
# A tibble: 9,955 × 4
Design ID Year_Type Height_cm
<chr> <dbl> <chr> <dbl>
1 1abc1abc8 45 2001_Height NA
2 1abc1abc8 45 2002_Height NA
3 1abc1abc8 45 2003_Height NA
4 1abc1abc8 45 2004_Height NA
5 1abc1abc8 45 2005_Height NA
6 1abc1abc216 44 2001_Height 547
7 1abc1abc216 44 2002_Height 622
8 1abc1abc216 44 2003_Height 715
9 1abc1abc216 44 2004_Height 716
10 1abc1abc216 44 2005_Height 817
# ℹ 9,945 more rowsseparate_wider_delim() examples
face_long_abc %>%
separate_wider_delim(cols = "Design",
delim = "abc",
names = c("Rep", "Treat", "Clone"))# A tibble: 9,955 × 6
Rep Treat Clone ID Year_Type Height_cm
<chr> <chr> <chr> <dbl> <chr> <dbl>
1 1 1 8 45 2001_Height NA
2 1 1 8 45 2002_Height NA
3 1 1 8 45 2003_Height NA
4 1 1 8 45 2004_Height NA
5 1 1 8 45 2005_Height NA
6 1 1 216 44 2001_Height 547
7 1 1 216 44 2002_Height 622
8 1 1 216 44 2003_Height 715
9 1 1 216 44 2004_Height 716
10 1 1 216 44 2005_Height 817
# ℹ 9,945 more rowsseparate_wider_delim() examples
# A tibble: 9,955 × 6
Rep Treat Clone ID Year_Type Height_cm
<dbl> <dbl> <dbl> <dbl> <chr> <dbl>
1 1 1 8 45 2001_Height NA
2 1 1 8 45 2002_Height NA
3 1 1 8 45 2003_Height NA
4 1 1 8 45 2004_Height NA
5 1 1 8 45 2005_Height NA
6 1 1 216 44 2001_Height 547
7 1 1 216 44 2002_Height 622
8 1 1 216 44 2003_Height 715
9 1 1 216 44 2004_Height 716
10 1 1 216 44 2005_Height 817
# ℹ 9,945 more rows# A tibble: 9,955 × 6
Rep Treat Clone ID Year Height_cm
<dbl> <dbl> <dbl> <dbl> <chr> <dbl>
1 1 1 8 45 2001 NA
2 1 1 8 45 2002 NA
3 1 1 8 45 2003 NA
4 1 1 8 45 2004 NA
5 1 1 8 45 2005 NA
6 1 1 216 44 2001 547
7 1 1 216 44 2002 622
8 1 1 216 44 2003 715
9 1 1 216 44 2004 716
10 1 1 216 44 2005 817
# ℹ 9,945 more rowsNext time
- More reshaping data with
tidyr