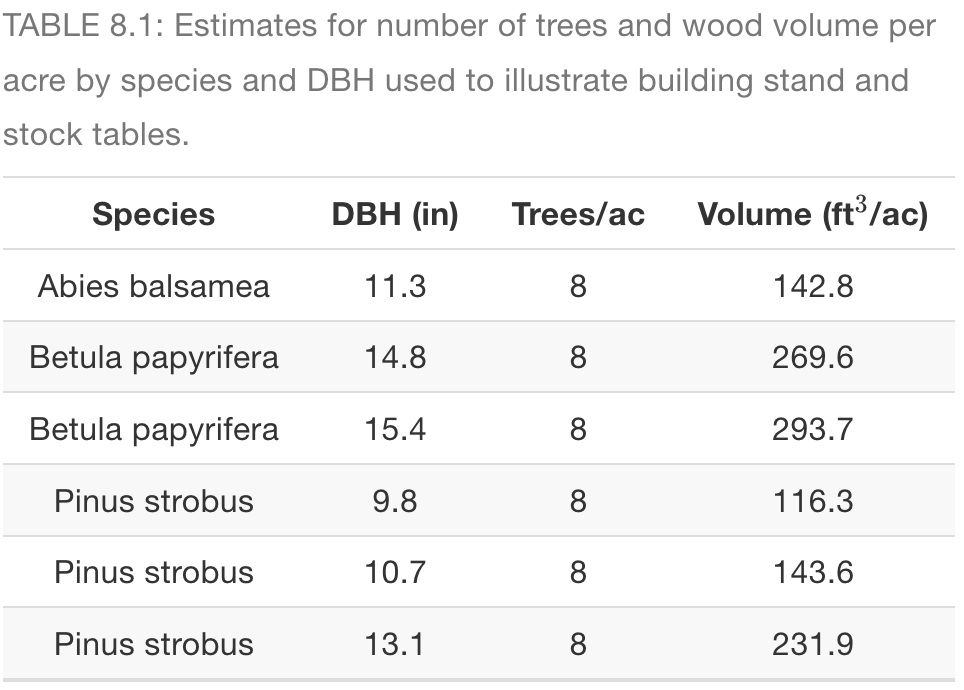
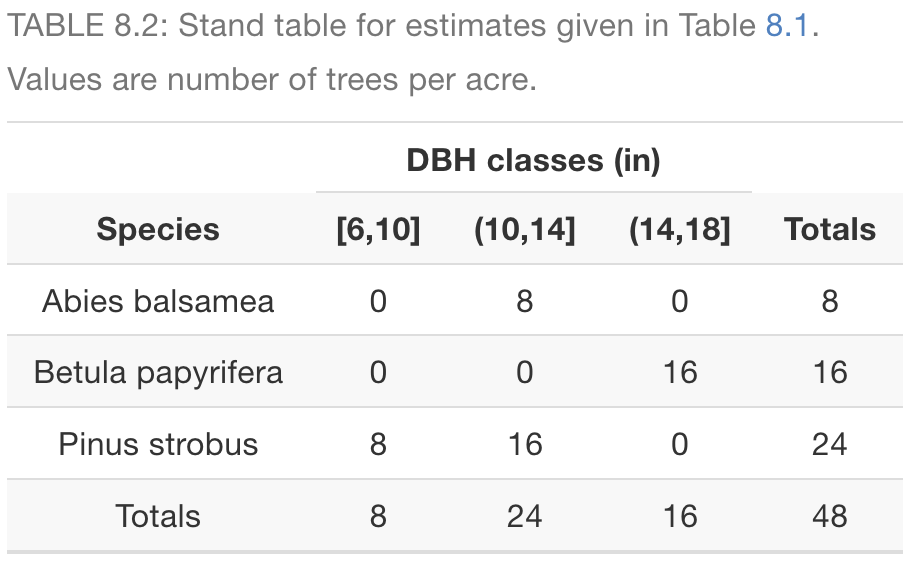
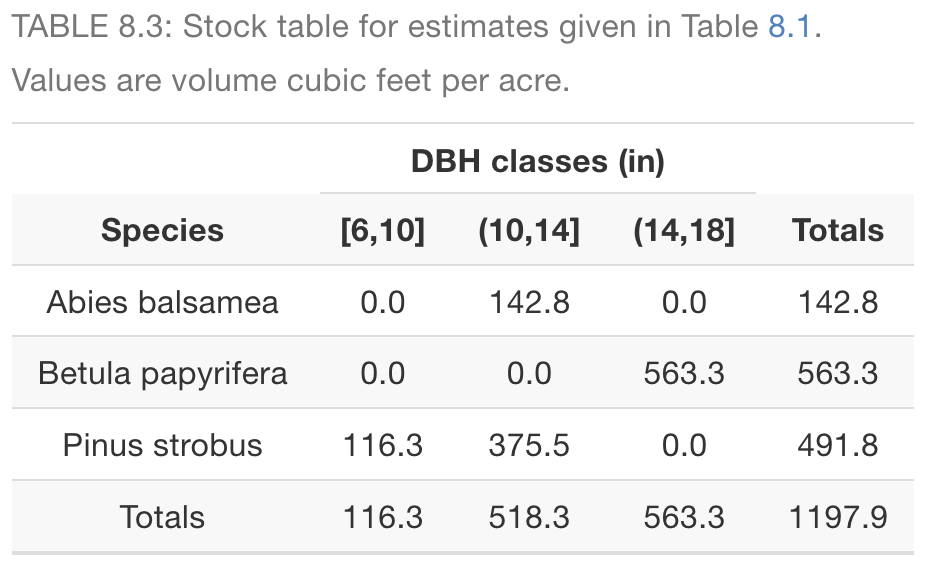
library(tidyverse)
face <- read_csv("../labs/datasets/FACE/FACE_aspen_core_growth.csv") %>%
select(Rep, Treat, Clone, ID = `ID #`,
contains(as.character(2001:2005)) & contains("Height"))
face# A tibble: 1,991 × 9
Rep Treat Clone ID `2001_Height` `2002_Height` `2003_Height`
<dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 1 1 8 45 NA NA NA
2 1 1 216 44 547 622 715
3 1 1 8 43 273 275 305
4 1 1 216 42 526 619 720
5 1 1 216 54 328 341 364
6 1 1 271 55 543 590 634
7 1 1 271 56 450 502 587
8 1 1 8 57 217 227 256
9 1 1 259 58 158 155 NA
10 1 1 271 59 230 241 260
# ℹ 1,981 more rows
# ℹ 2 more variables: `2004_Height` <dbl>, `2005_Height` <dbl>