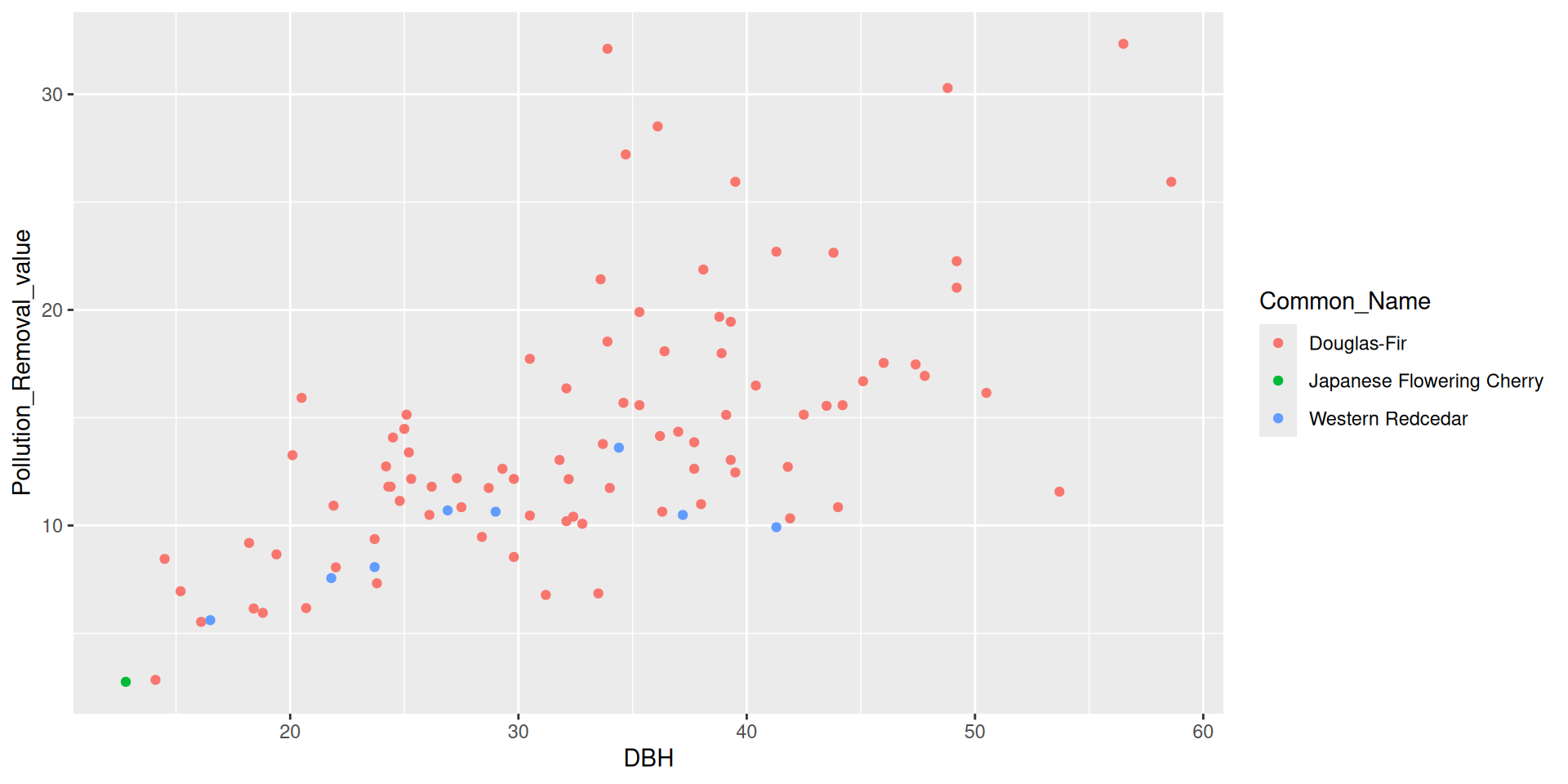
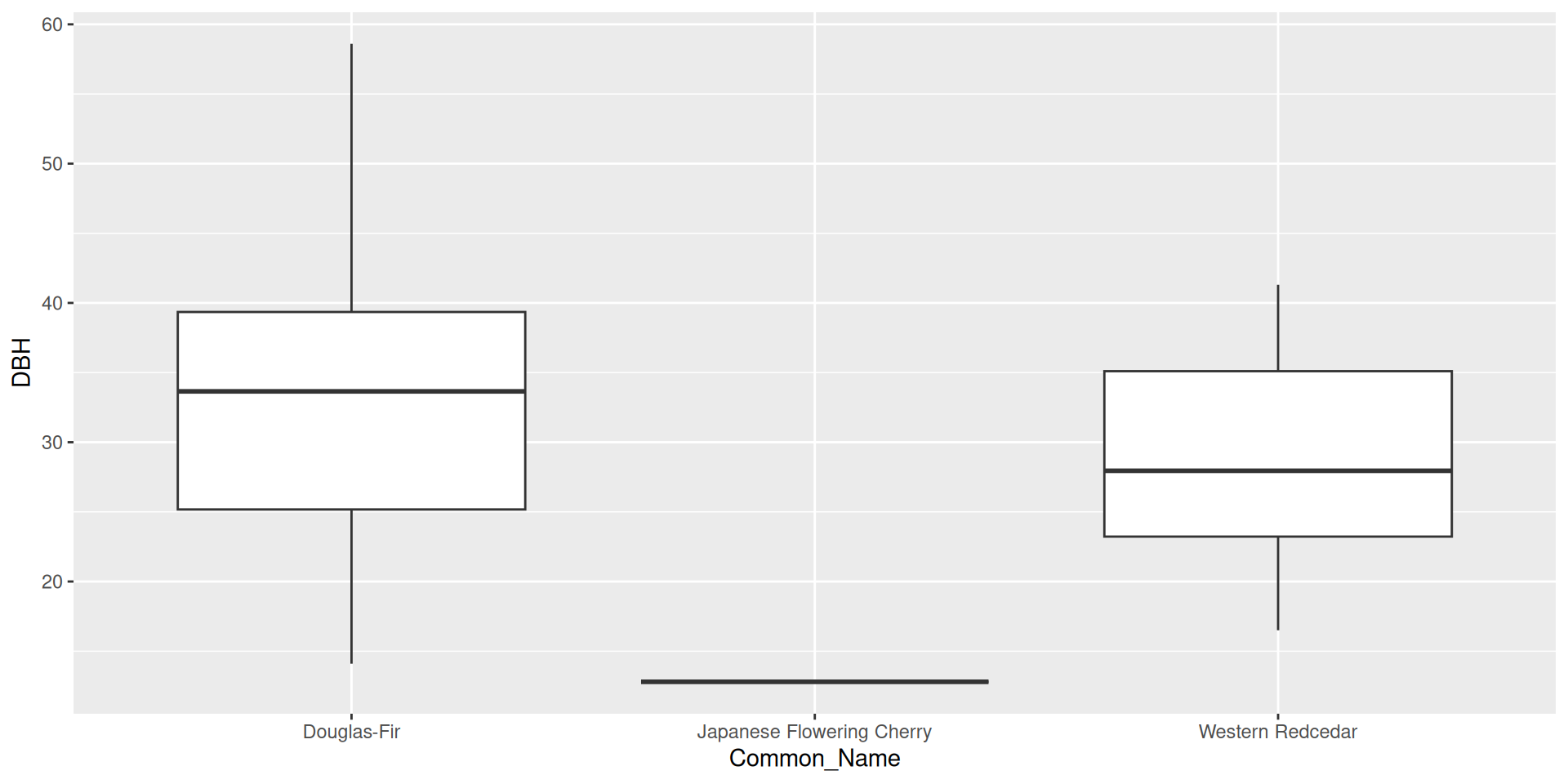
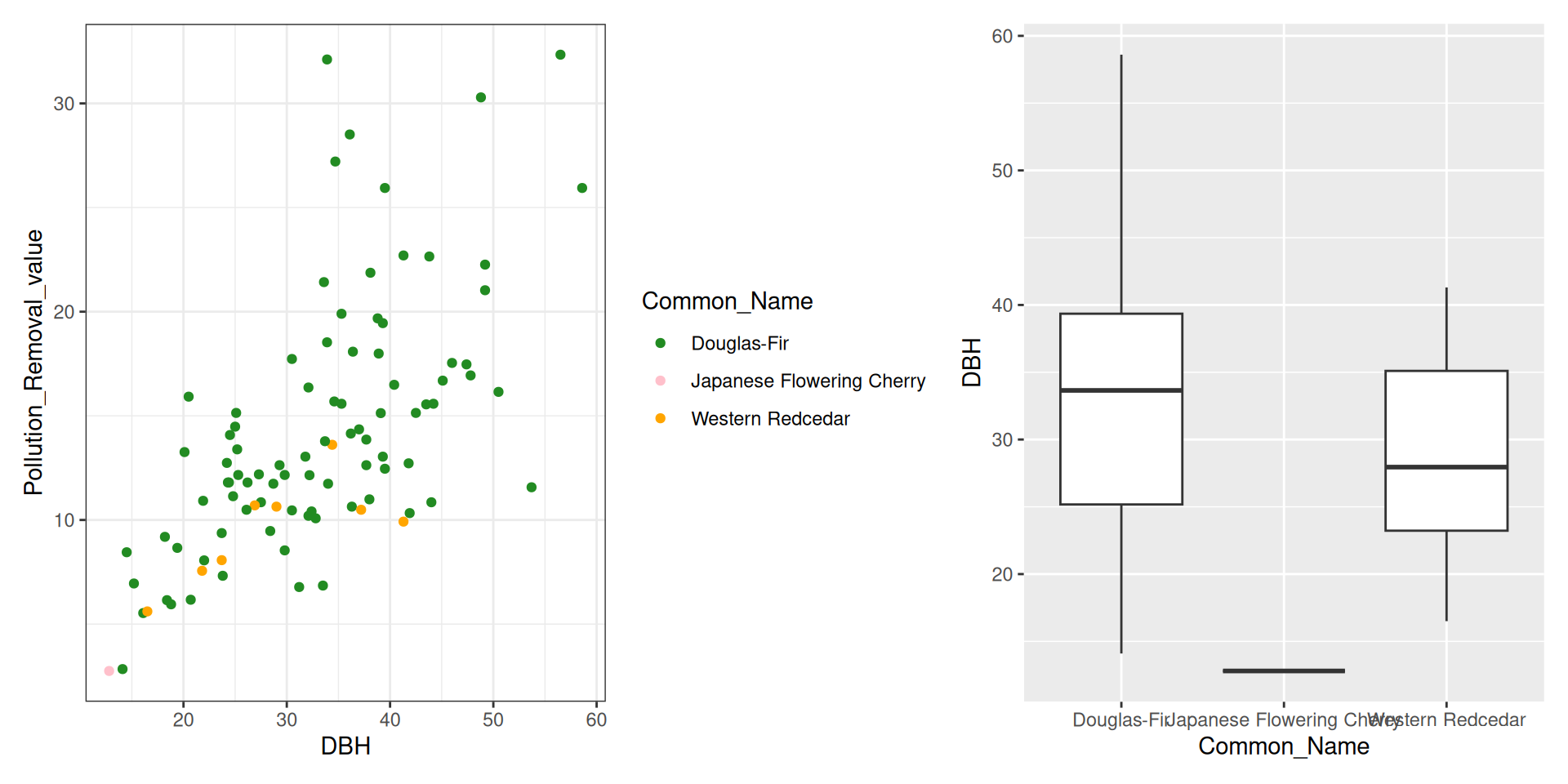
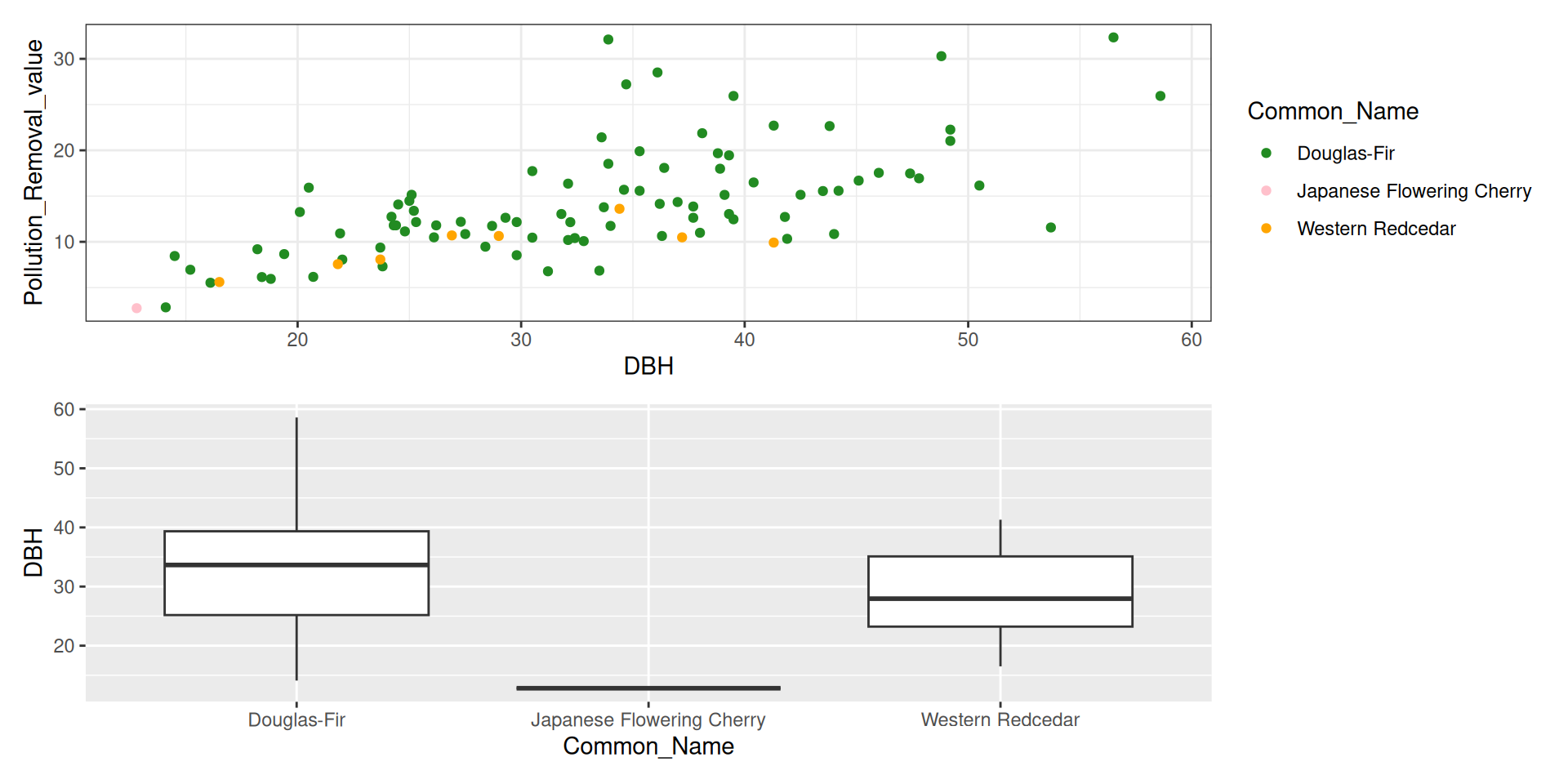
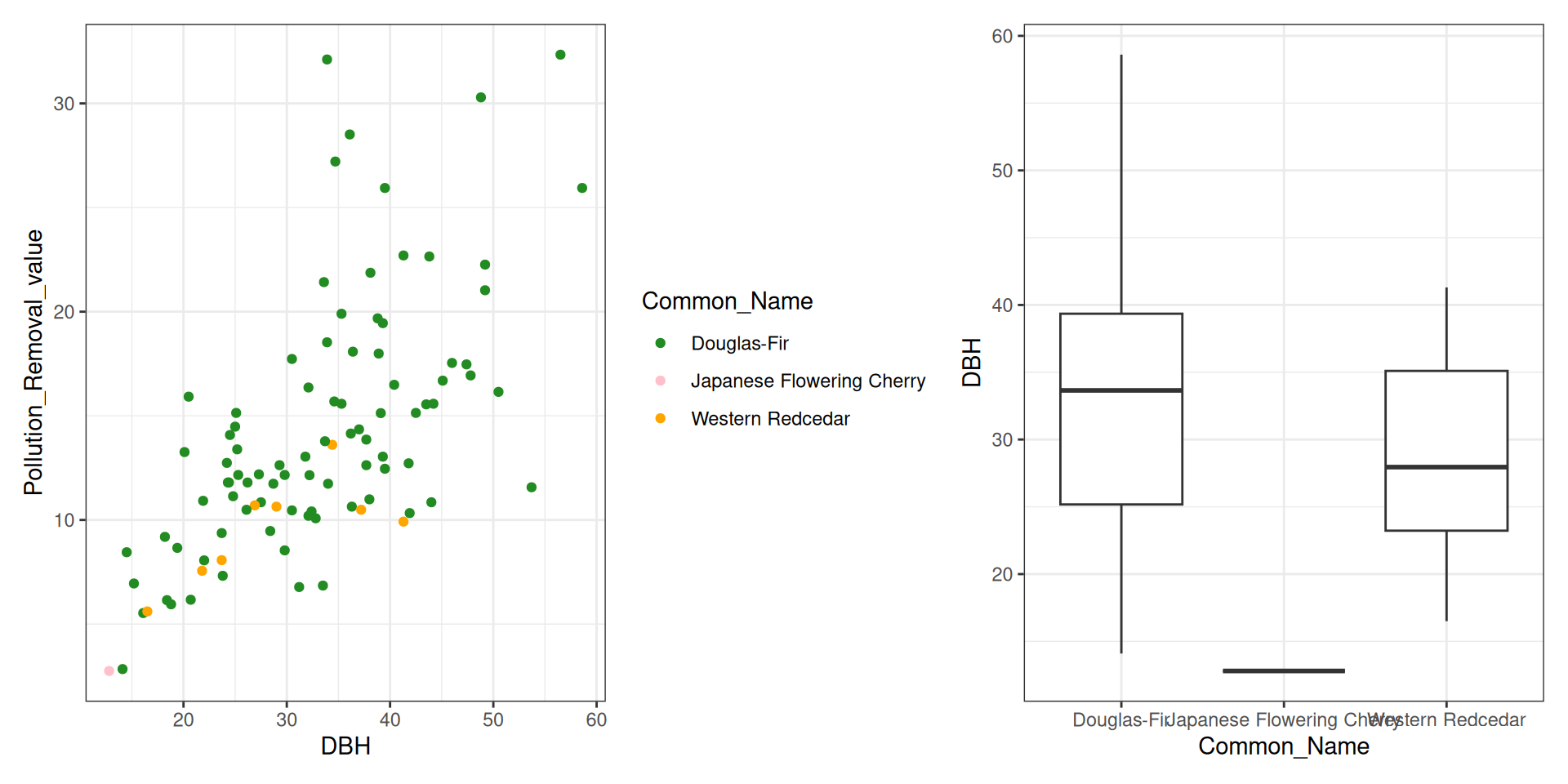
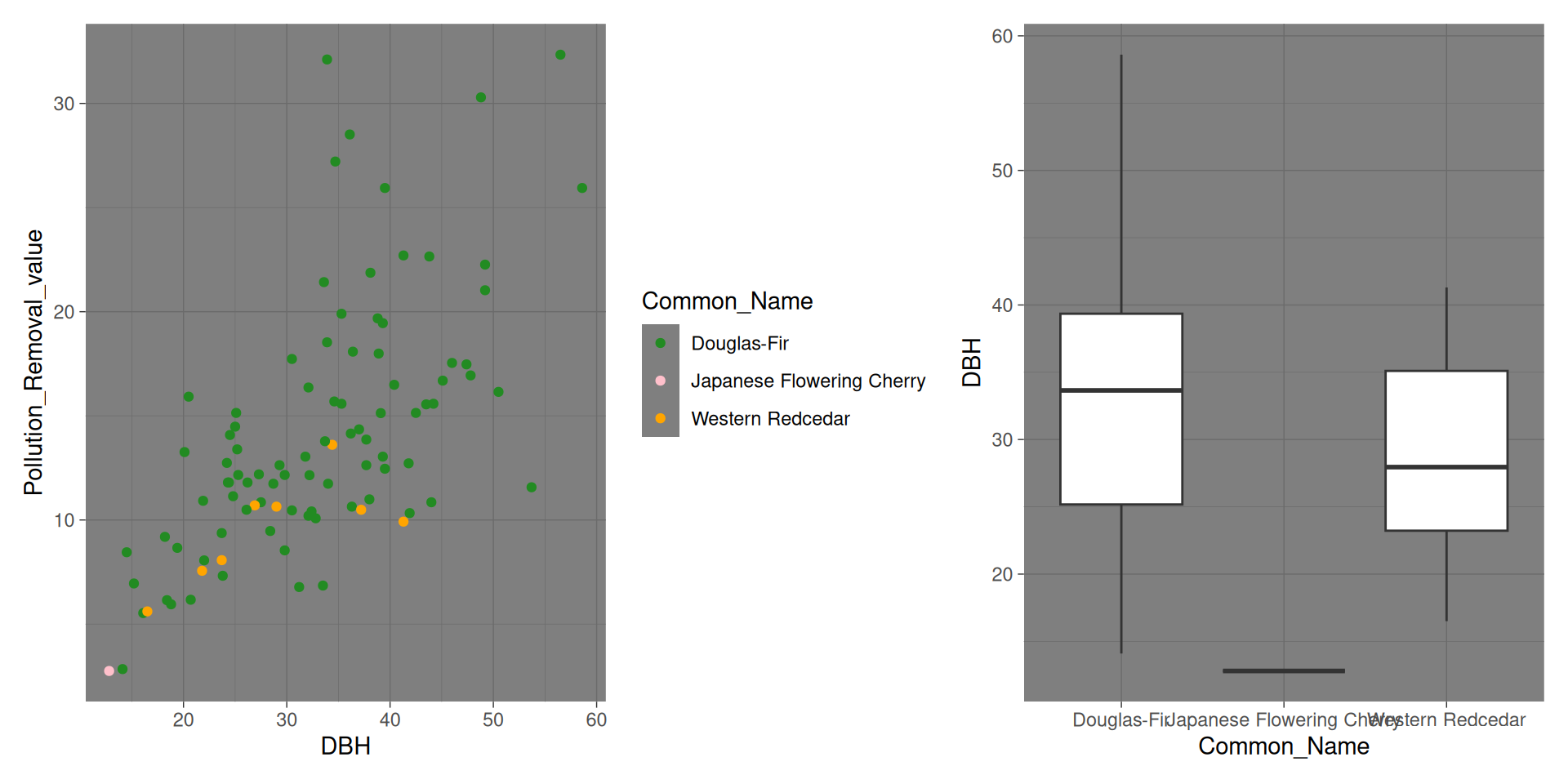
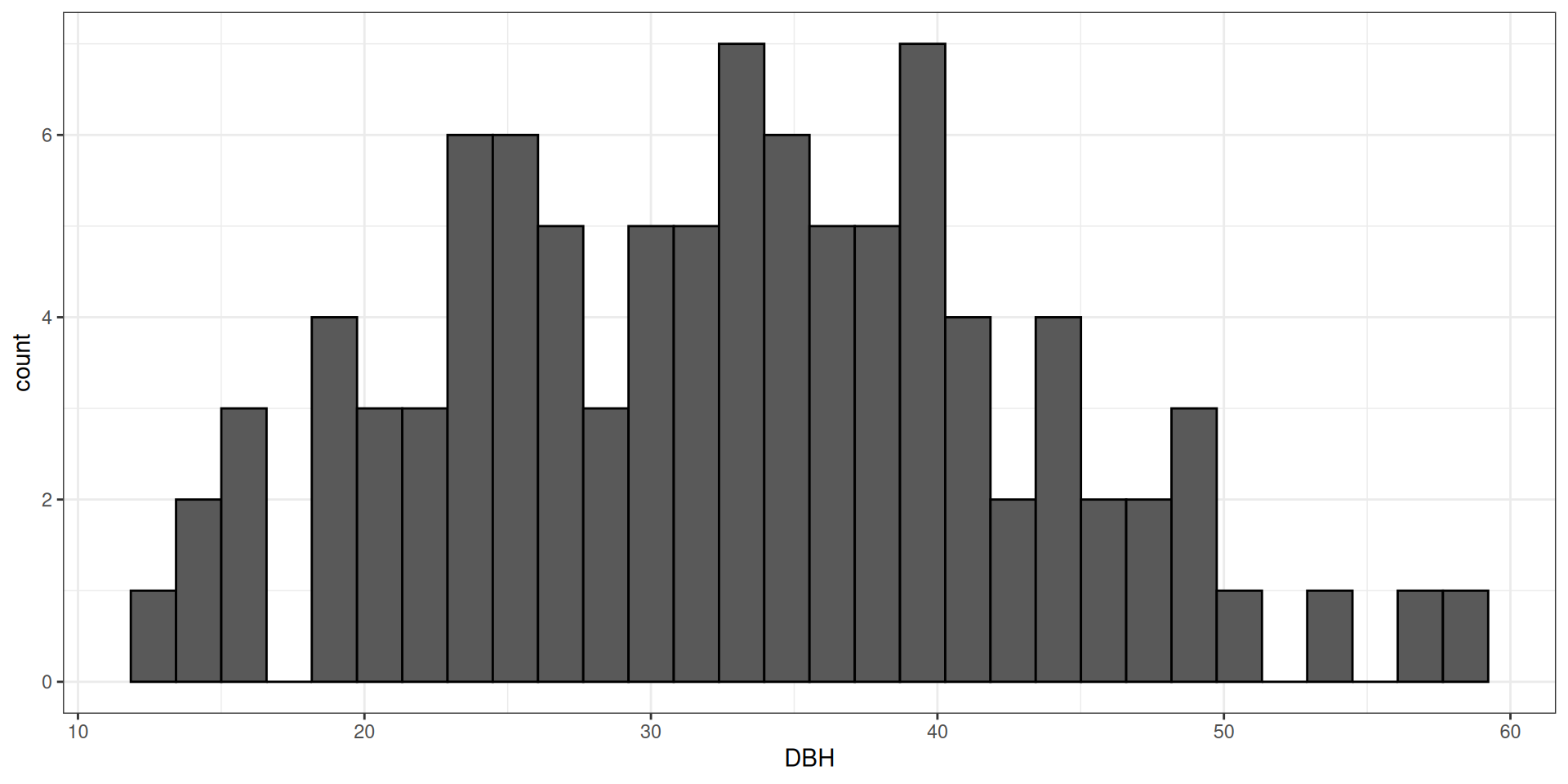
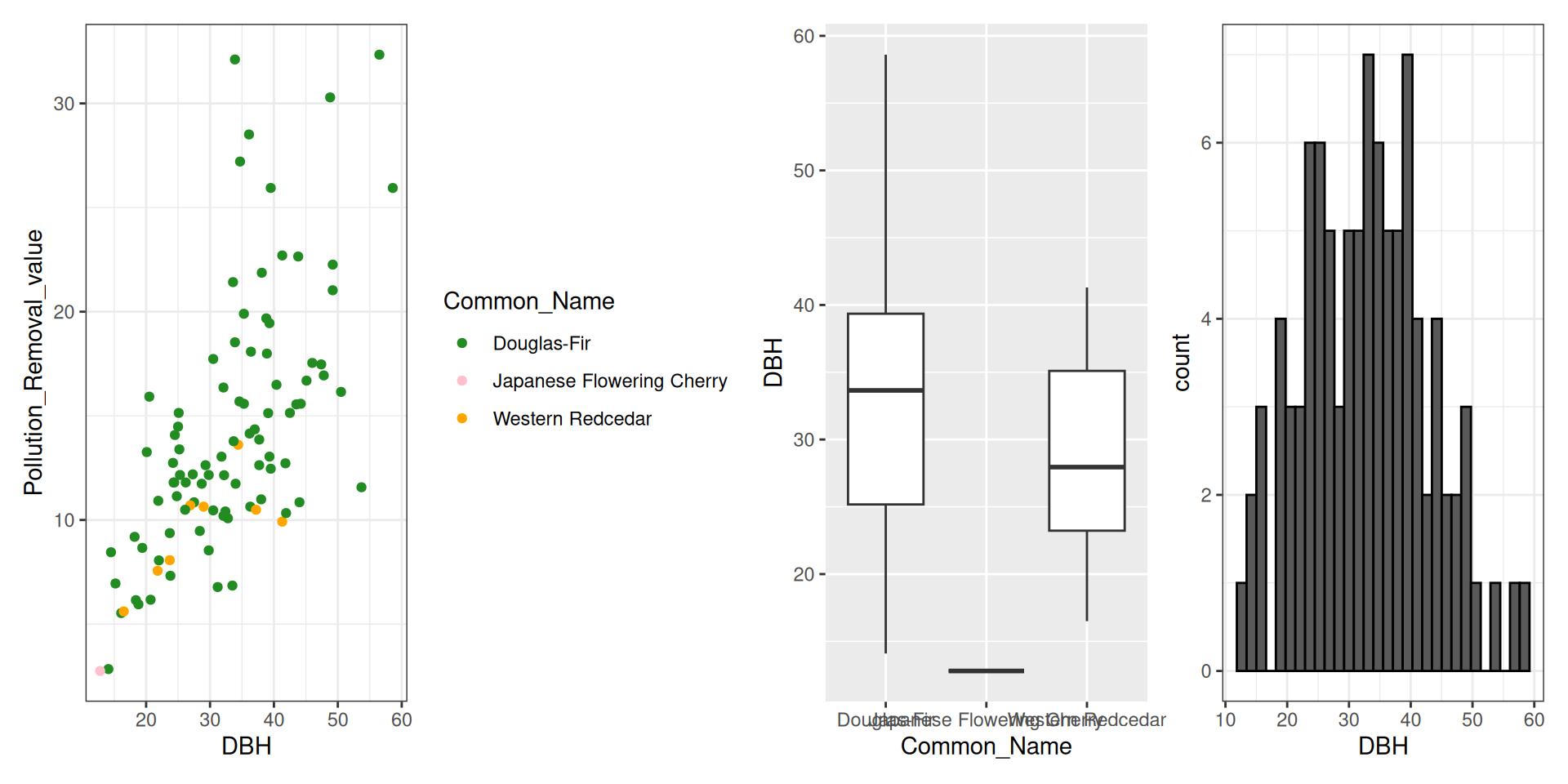
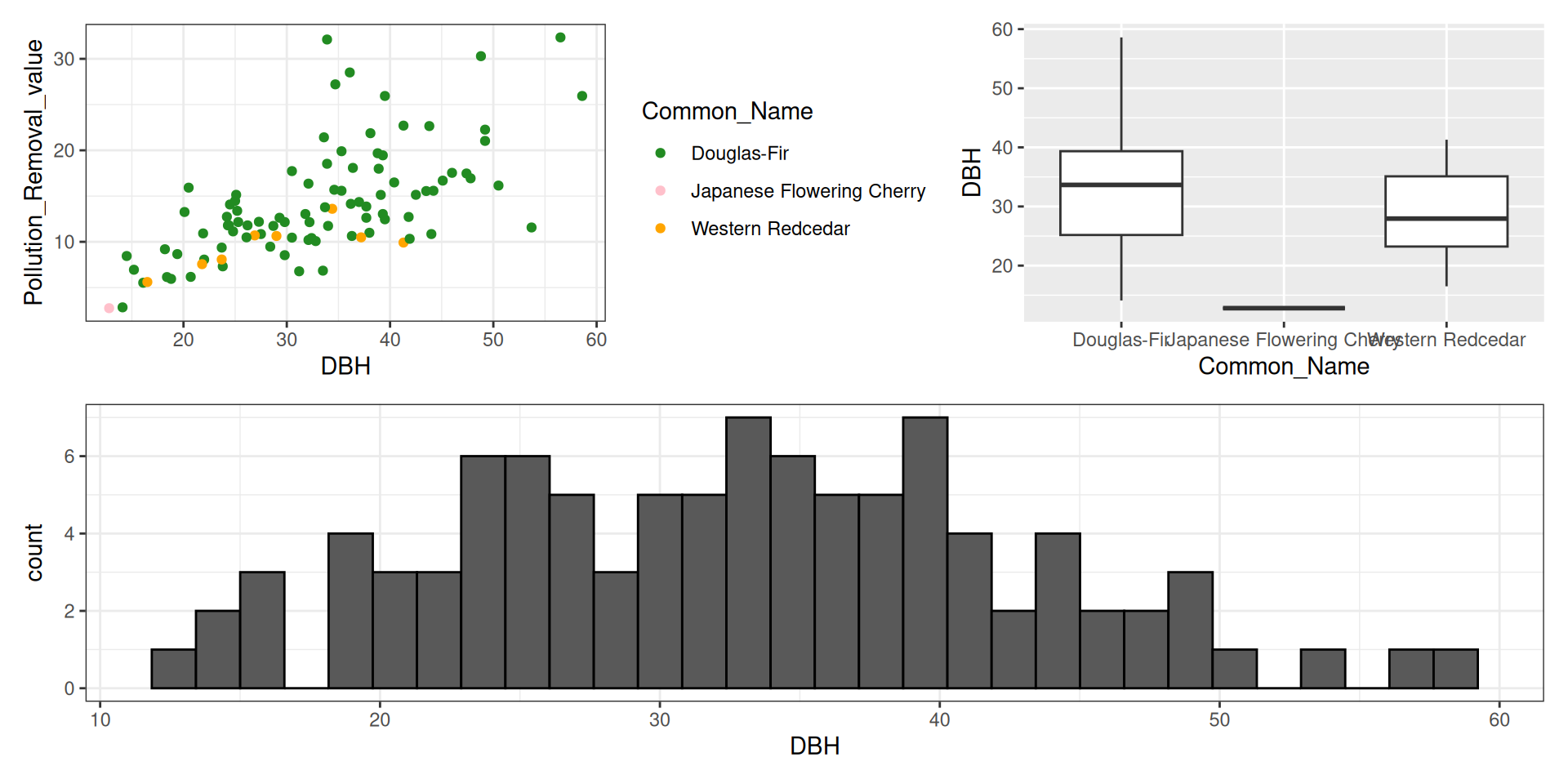
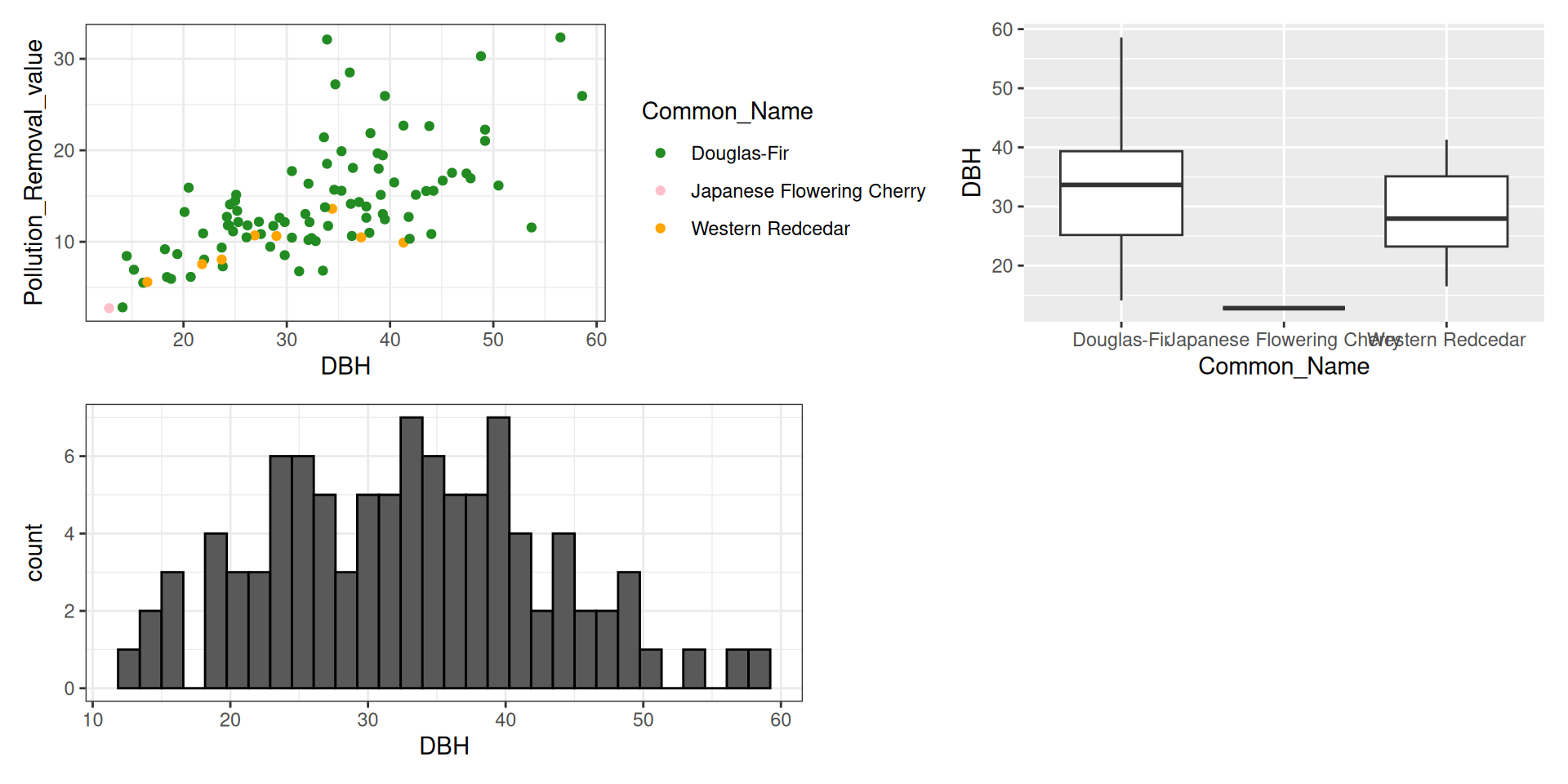
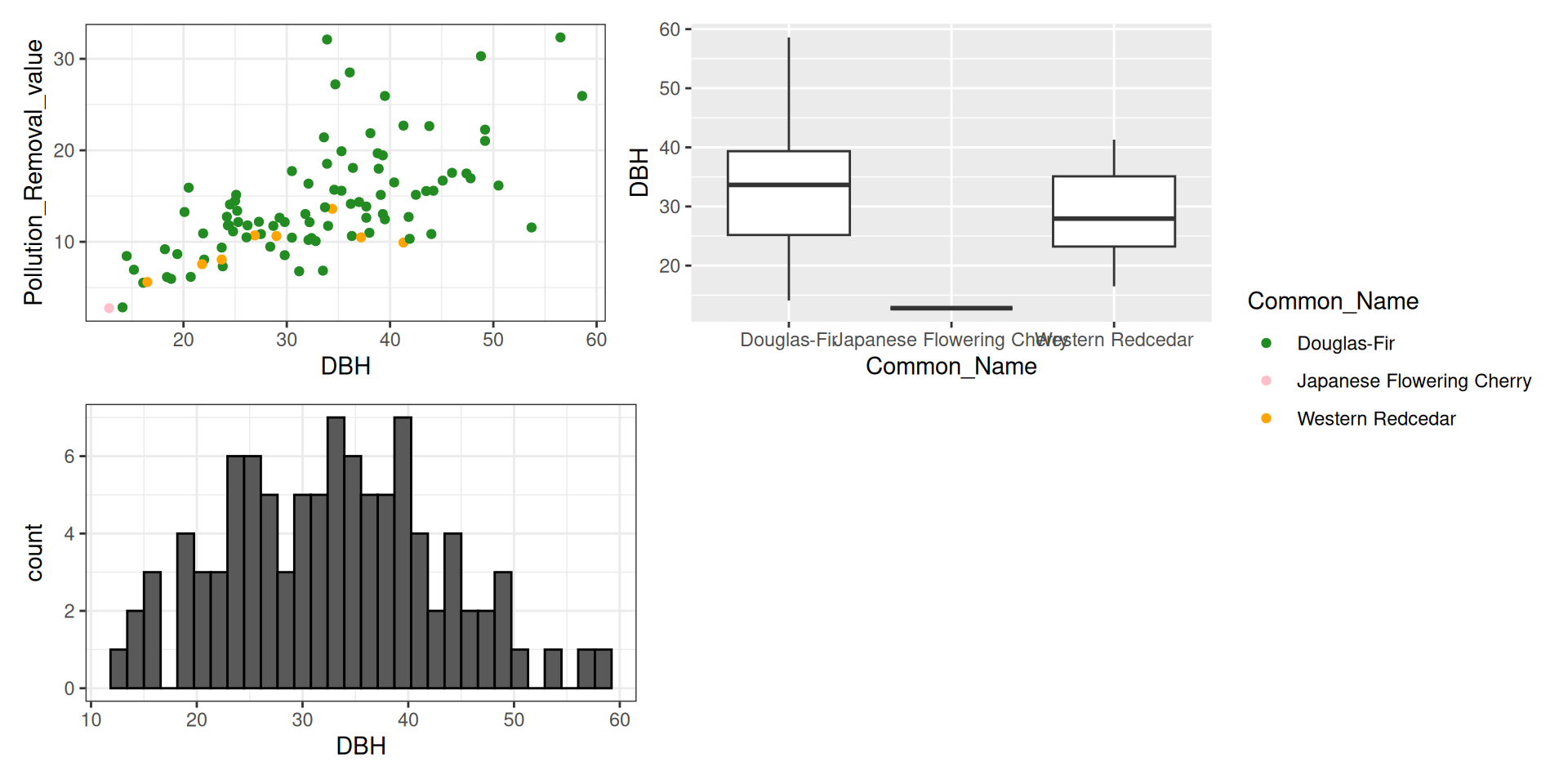
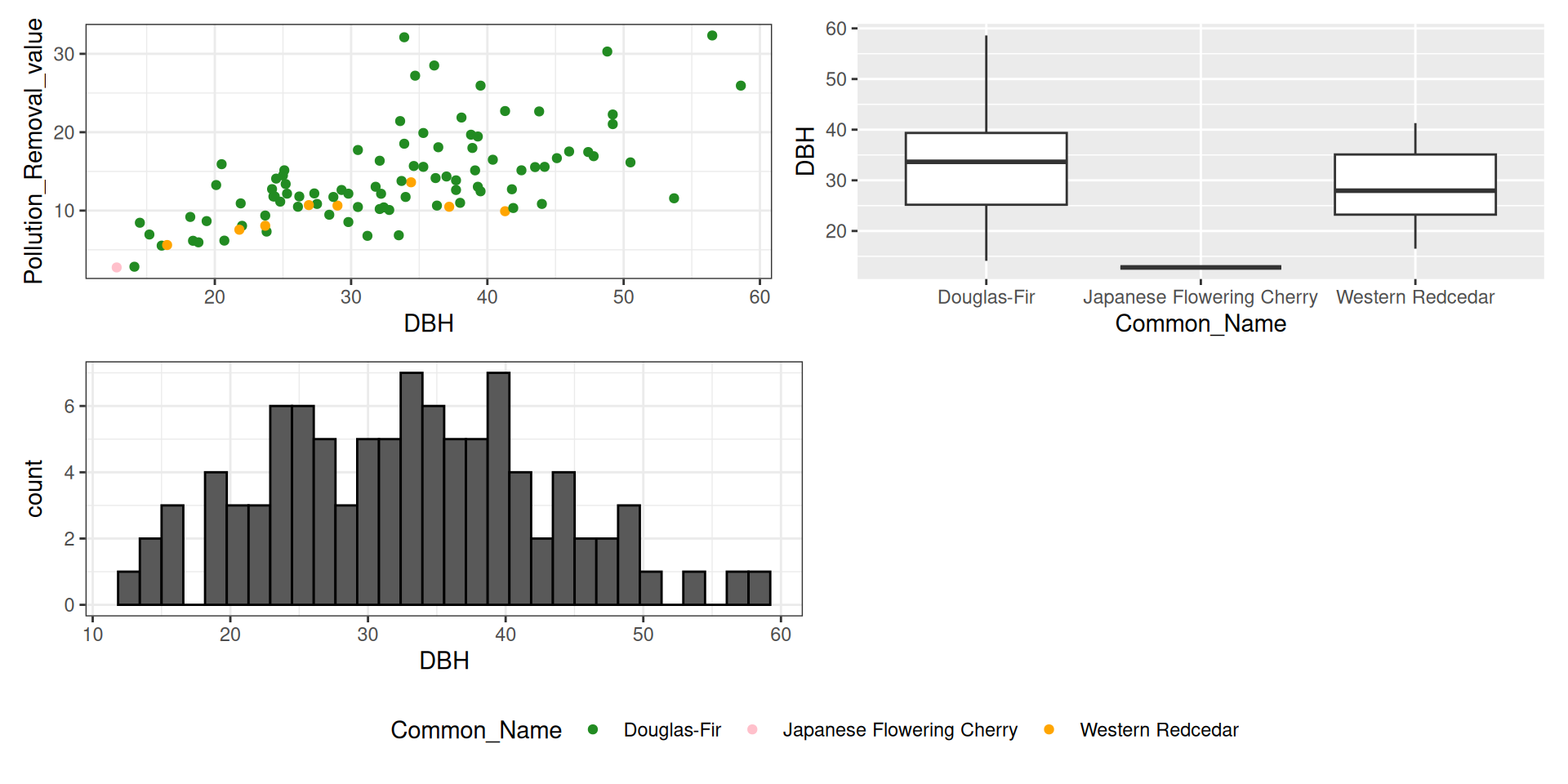
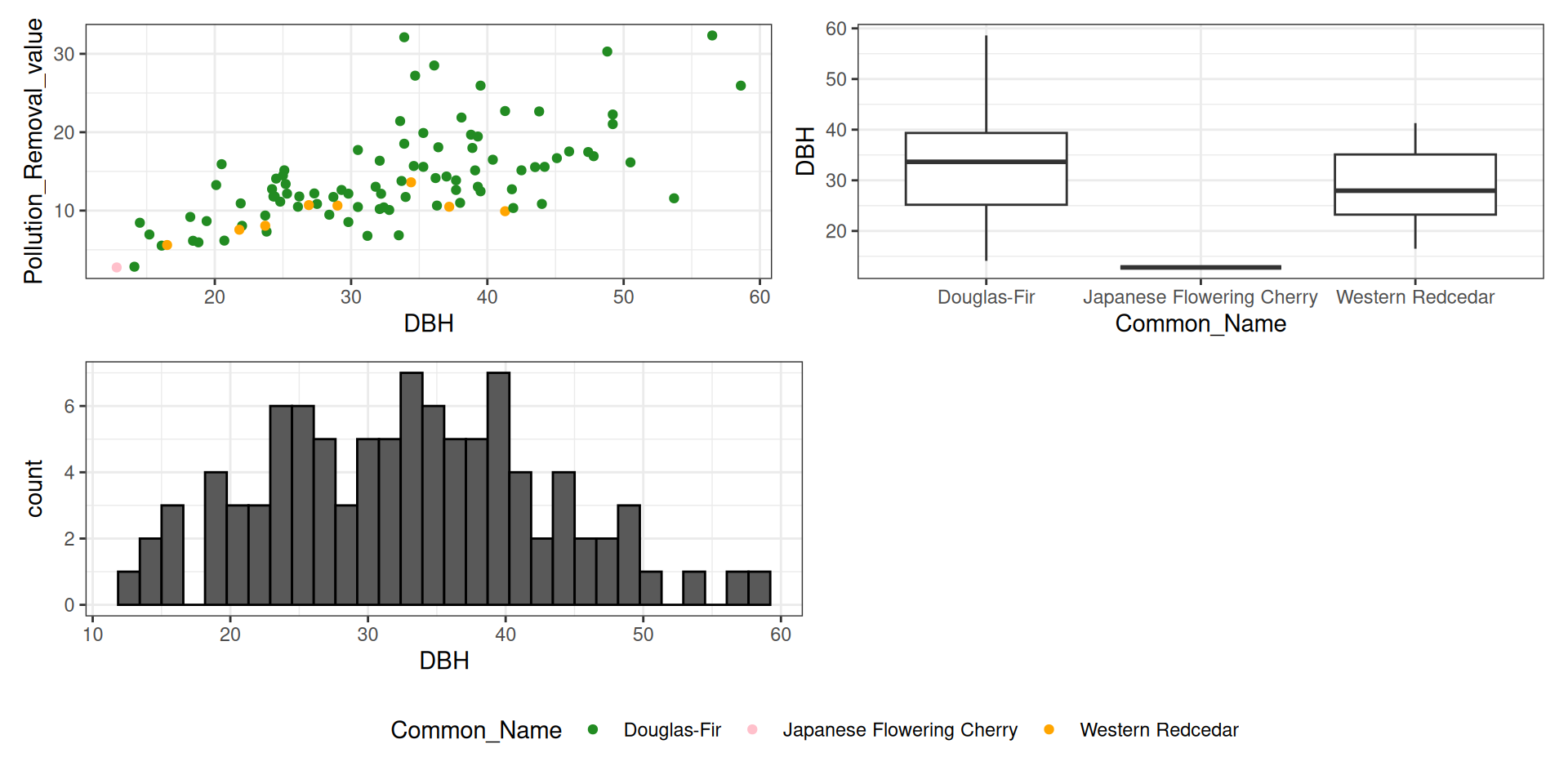
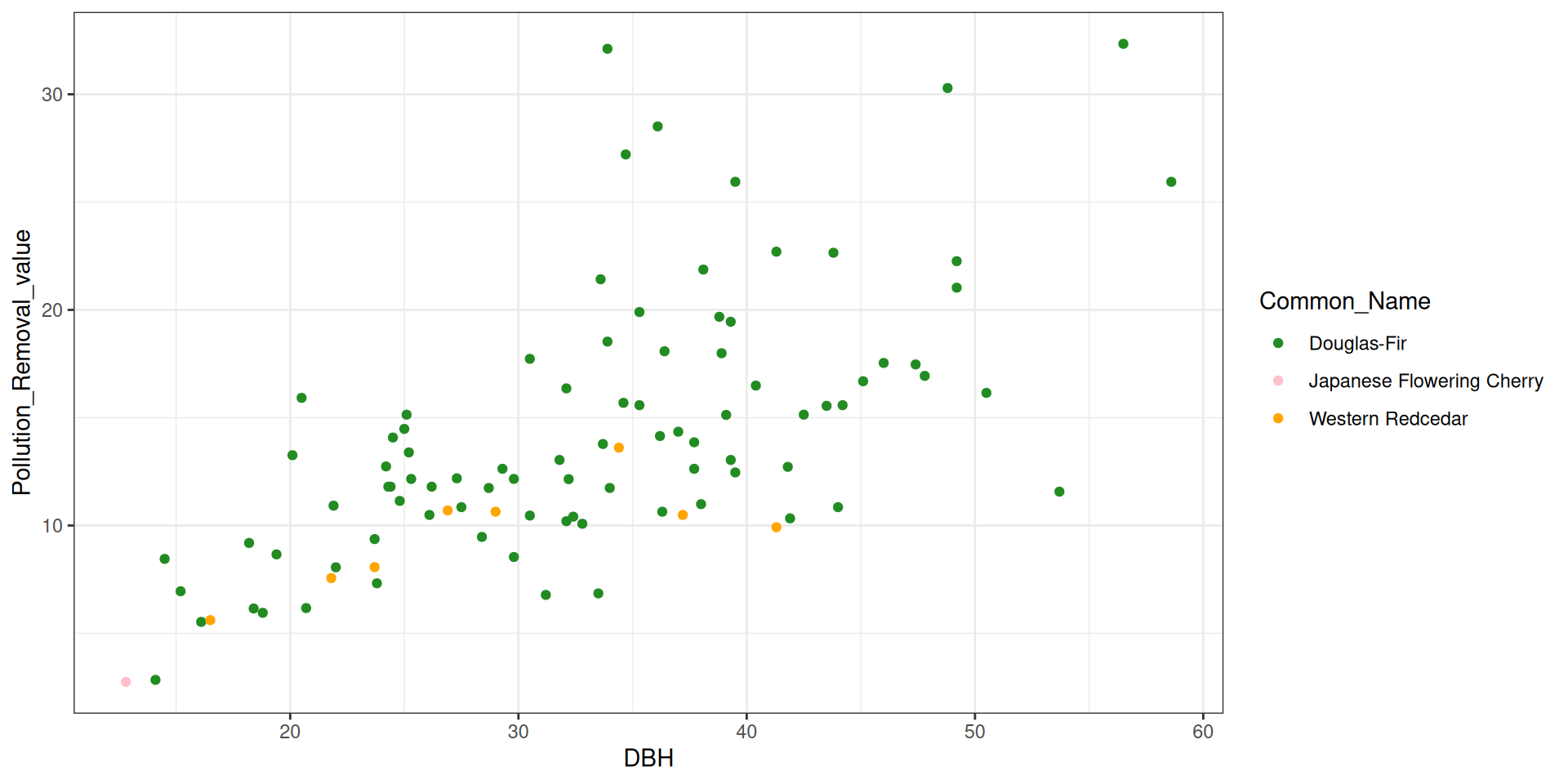
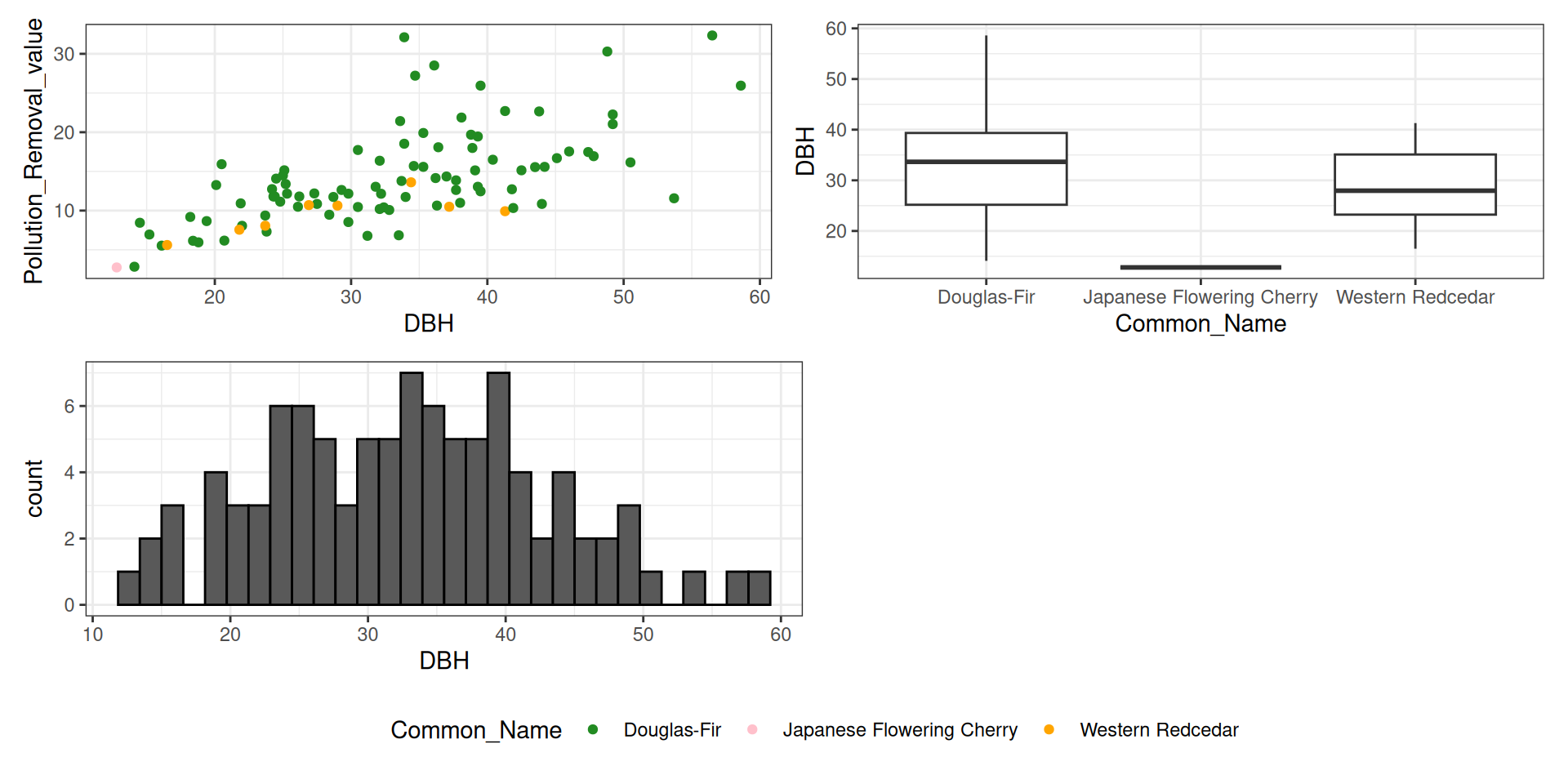
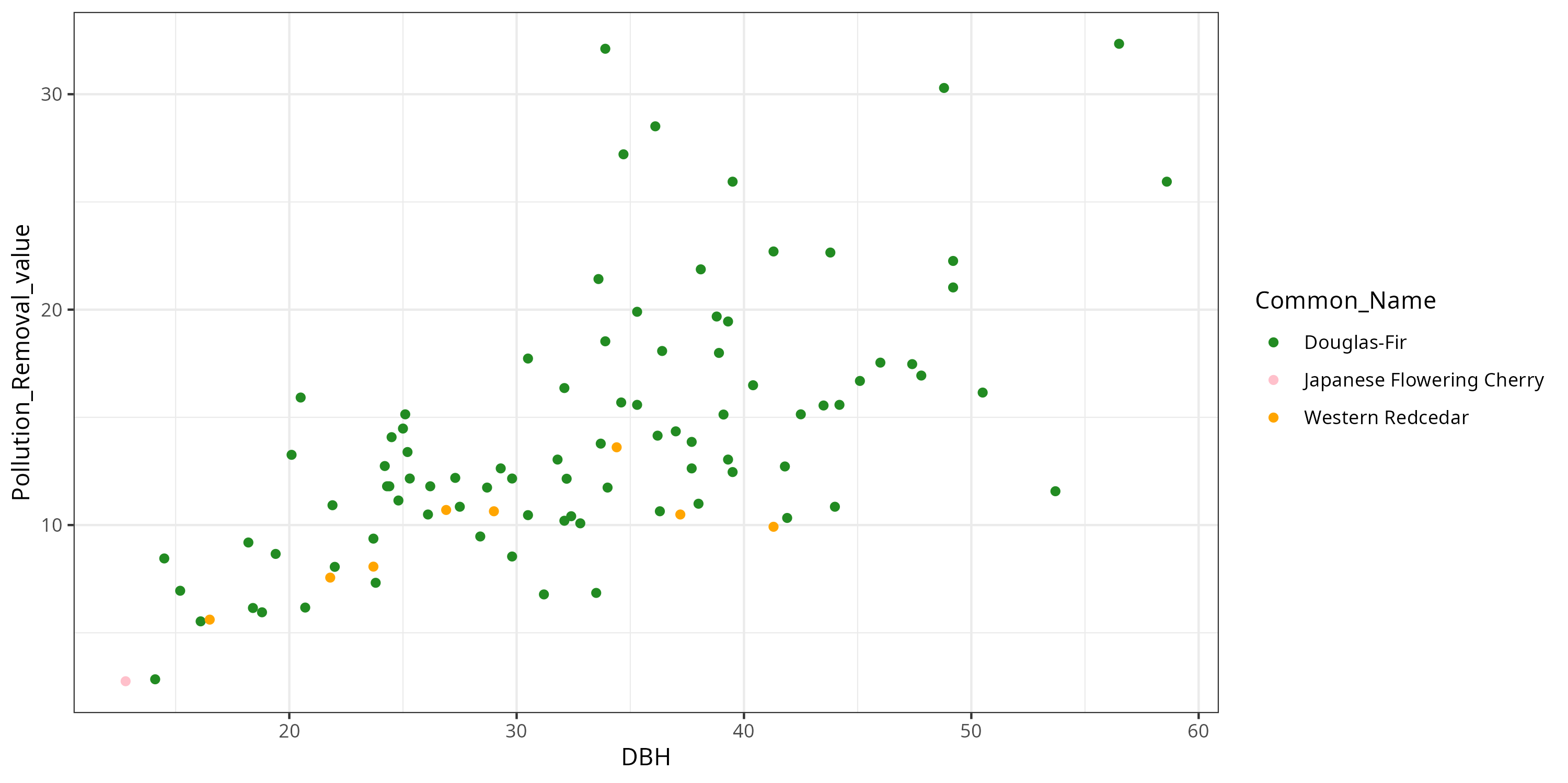
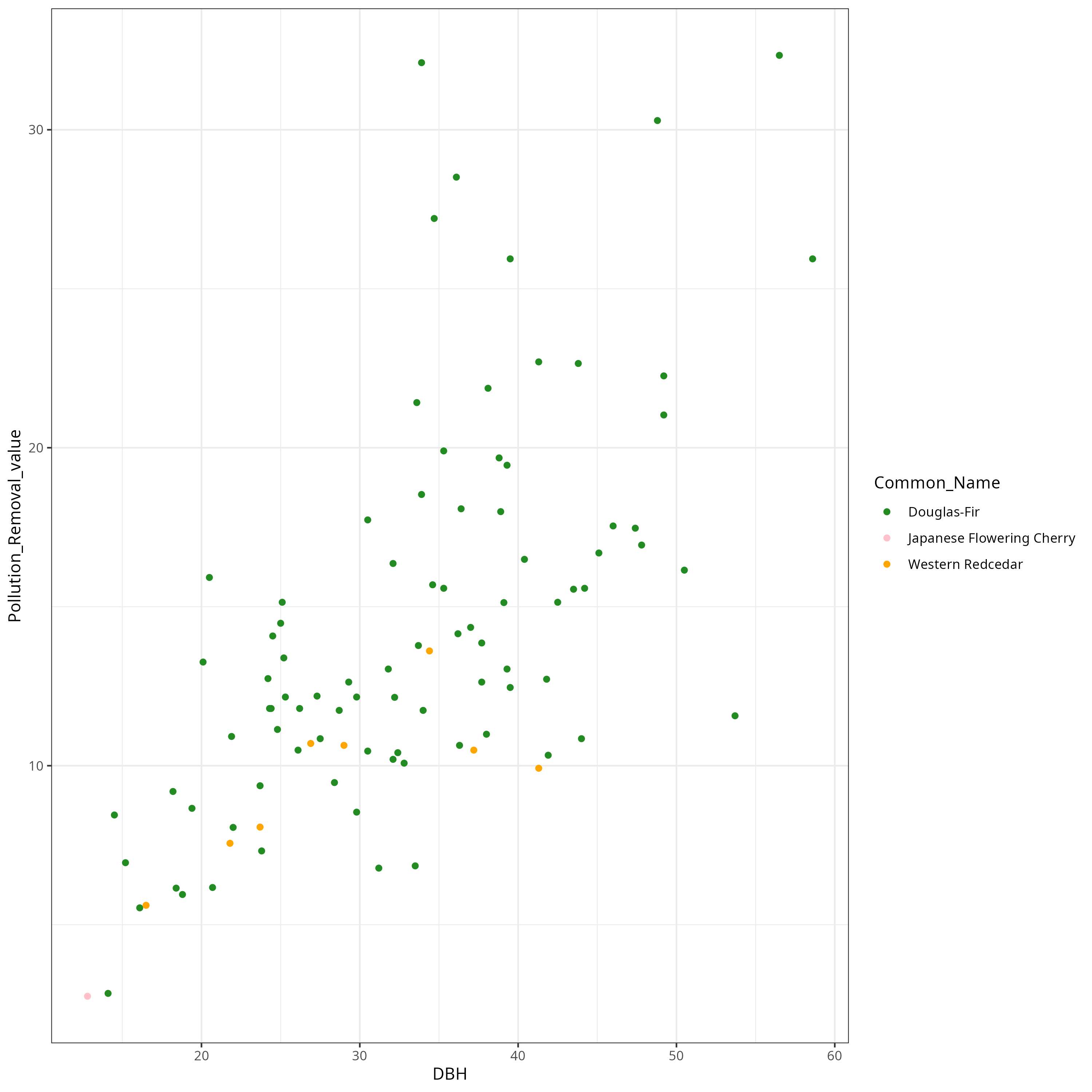
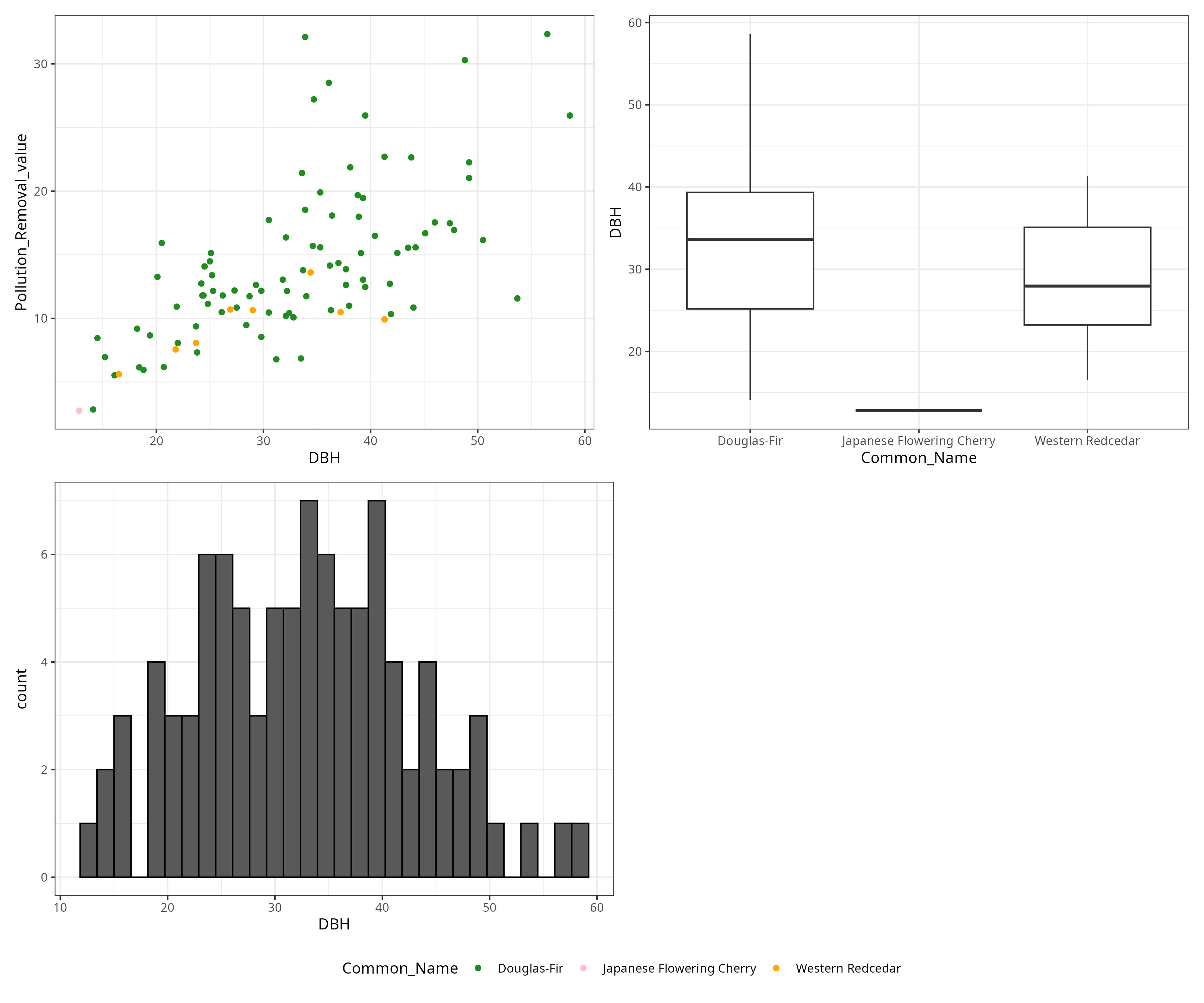
library(tidyverse)
library(pdxTrees)
dat <- get_pdxTrees_parks(park = c("Woodstock Park")) %>%
filter(Common_Name %in% c("Douglas-Fir",
"Japanese Flowering Cherry",
"Western Redcedar"))
dat
# A tibble: 97 × 34
Longitude Latitude UserID Genus Family DBH Inventory_Date Species
<dbl> <dbl> <chr> <chr> <chr> <dbl> <dttm> <chr>
1 -123. 45.5 18381 Pseudotsu… Pinac… 41.3 2019-07-20 00:00:00 PSME
2 -123. 45.5 18386 Pseudotsu… Pinac… 34.7 2019-07-20 00:00:00 PSME
3 -123. 45.5 18415 Pseudotsu… Pinac… 36.3 2019-07-20 00:00:00 PSME
4 -123. 45.5 18417 Pseudotsu… Pinac… 32.4 2019-07-20 00:00:00 PSME
5 -123. 45.5 18420 Pseudotsu… Pinac… 18.2 2019-07-20 00:00:00 PSME
6 -123. 45.5 18423 Thuja Cupre… 34.4 2019-07-20 00:00:00 THPL
7 -123. 45.5 18486 Pseudotsu… Pinac… 38 2019-07-20 00:00:00 PSME
8 -123. 45.5 18488 Pseudotsu… Pinac… 31.2 2019-07-20 00:00:00 PSME
9 -123. 45.5 18489 Pseudotsu… Pinac… 32.2 2019-07-20 00:00:00 PSME
10 -123. 45.5 18490 Pseudotsu… Pinac… 49.2 2019-07-20 00:00:00 PSME
# ℹ 87 more rows
# ℹ 26 more variables: Common_Name <chr>, Condition <chr>, Tree_Height <dbl>,
# Crown_Width_NS <dbl>, Crown_Width_EW <dbl>, Crown_Base_Height <dbl>,
# Collected_By <chr>, Park <chr>, Scientific_Name <chr>,
# Functional_Type <chr>, Mature_Size <fct>, Native <chr>, Edible <chr>,
# Nuisance <chr>, Structural_Value <dbl>, Carbon_Storage_lb <dbl>,
# Carbon_Storage_value <dbl>, Carbon_Sequestration_lb <dbl>, …