[1] "/home/grayson/courses/FOR128/FOR128.github.io/slides"Reading and Writing Data
Agenda
- Quiz 1 review
- Navigating your computer
- Reading and writing data
Quiz Review
Navigating your computer
Recall your computer’s file system
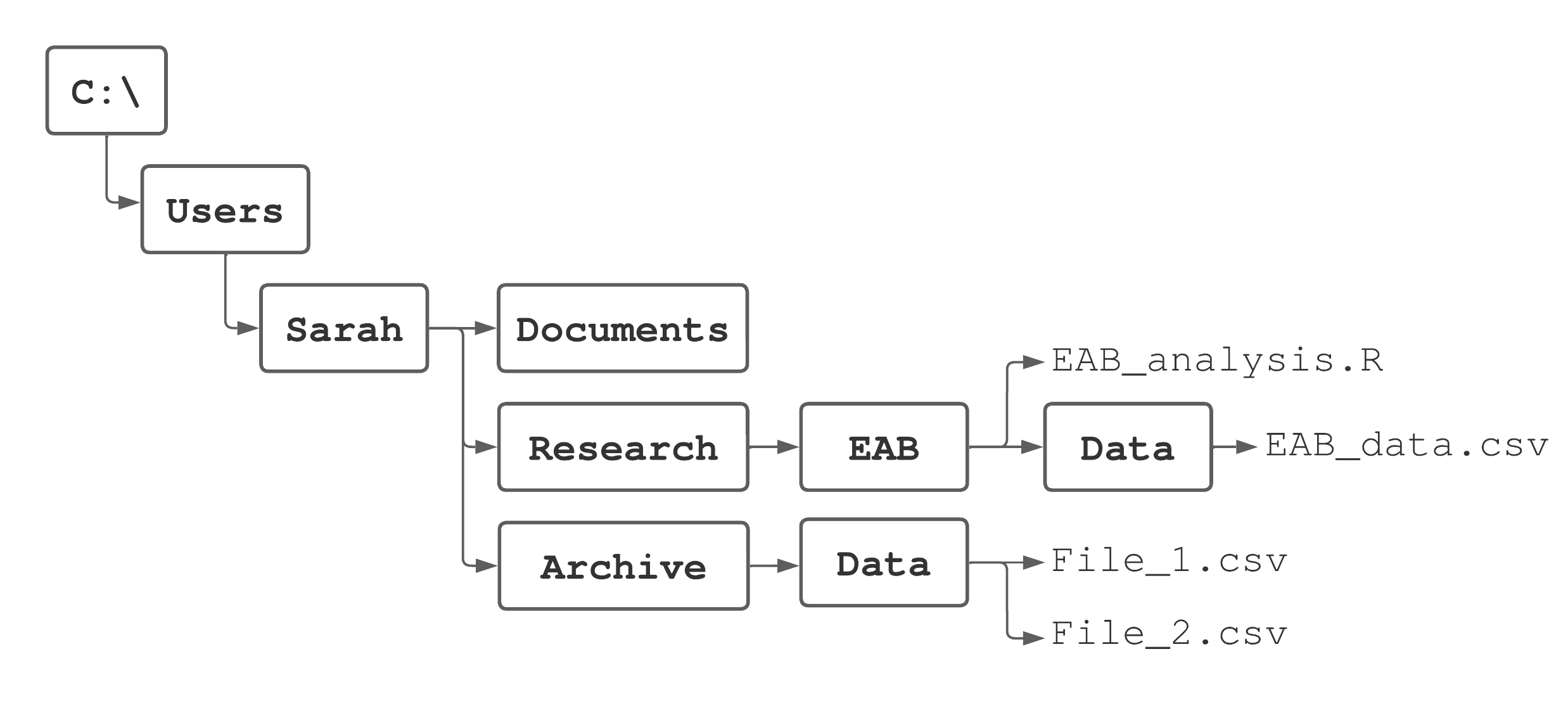
What is the root directory? 01:00
01:00 
What is the root directory?

C:/
What is the home directory? 01:00
01:00 
What is the home directory?

C:/Users/Sarah
How do we specify the absolute path to File_1.csv? 01:00
01:00 
How do we specify the absolute path to File_1.csv?

C:/Users/Sarah/Archive/Data/File_1.csv
How do we specify the relative path to File_1.csv, if Sarah’s home directory is her working directory? 01:00
01:00 
How do we specify the relative path to File_1.csv, if Sarah’s home directory is her working directory?

Archive/Data/File_1.csv
How do we specify the relative path to File_1.csv, if Sarah’s home directory is C:/Users/Sarah/Documents? 01:00
01:00 
How do we specify the relative path to File_1.csv, if Sarah’s home directory is C:/Users/Sarah/Documents?

../Archive/Data/File_1.csv
What’s going on with those two dots? (../) 01:00
01:00 Using
..in a file path means “go back a level”Example: Say I want to access a file on my desktop:
/home/grayson/Desktop/important_file.pdf
But, my working directory is as follows:
/home/grayson/Documents
Q: What is the relative path to the important file on my desktop?
A:
../Desktop/important_file.pdf
Now consider a different computer 01:00
01:00 We have an absolute path to
lab_01.qmd:/home/elliot/Documents/for128/lab_01.qmd
Say Elliot is using the following working directory:
/home/elliot/research/spatio_temporal
Q1: What operating system might Elliot be using?
Q2: What is the relative path to
lab_01.qmd?A1: macOS or Linux
A2:
../../for128/lab_01.qmd
Reading and writing data
Reading data
In this course, we’ll focus on reading 2-dimensional data (things that look like spreadsheets) into R.
Reading data
Reading data
Reading data
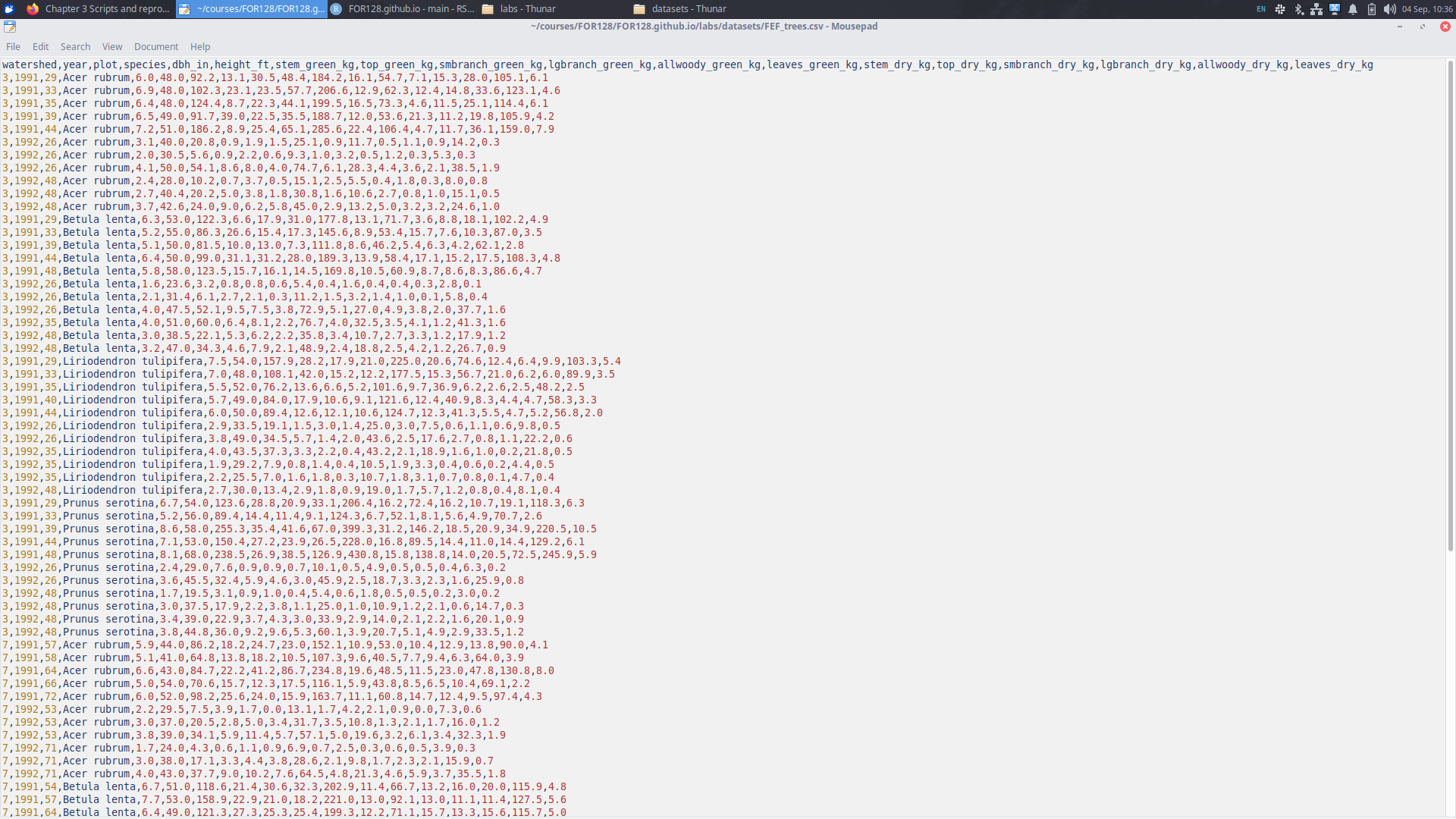
R includes a variety of functions for reading data efficiently. One of the most common is read.csv().
Reading data
R includes a variety of functions for reading data efficiently. One of the most common is read.csv().
To read data, we first may want to check our working directory:
Reading data
R includes a variety of functions for reading data efficiently. One of the most common is read.csv().
To read data, we first may want to check our working directory:
Then we can find the datasets folder. I’ve kept mine with my labs:
[1] "data_lab9" "data_lab9.zip" "datasets"
[4] "FEF_trees.csv" "lab_01_solutions.pdf" "lab_01_solutions.qmd"
[7] "lab_01.qmd" "lab_02.qmd" "lab_03_example.R"
[10] "lab_03.html" "lab_03.qmd" "lab_04.qmd"
[13] "lab_05.qmd" "lab_06.qmd" "lab_07_web.html"
[16] "lab_07_web.qmd" "lab_07.qmd" "lab_08_web.html"
[19] "lab_08_web.qmd" "lab_08.qmd" "lab_09_web.html"
[22] "lab_09_web.qmd" "lab_09.qmd" "lab_10_web_files"
[25] "lab_10_web.html" "lab_10_web.qmd" "lab_10.qmd"
[28] "lab_11.qmd" Reading data
R includes a variety of functions for reading data efficiently. One of the most common is read.csv().
To read data, we first may want to check our working directory:
Then we can find the datasets folder. I’ve kept mine with my labs:
[1] "data_lab9" "data_lab9.zip" "datasets"
[4] "FEF_trees.csv" "lab_01_solutions.pdf" "lab_01_solutions.qmd"
[7] "lab_01.qmd" "lab_02.qmd" "lab_03_example.R"
[10] "lab_03.html" "lab_03.qmd" "lab_04.qmd"
[13] "lab_05.qmd" "lab_06.qmd" "lab_07_web.html"
[16] "lab_07_web.qmd" "lab_07.qmd" "lab_08_web.html"
[19] "lab_08_web.qmd" "lab_08.qmd" "lab_09_web.html"
[22] "lab_09_web.qmd" "lab_09.qmd" "lab_10_web_files"
[25] "lab_10_web.html" "lab_10_web.qmd" "lab_10.qmd"
[28] "lab_11.qmd" Now, given our working directory and datasets location, we specify a relative path and load in the data:
Reading data
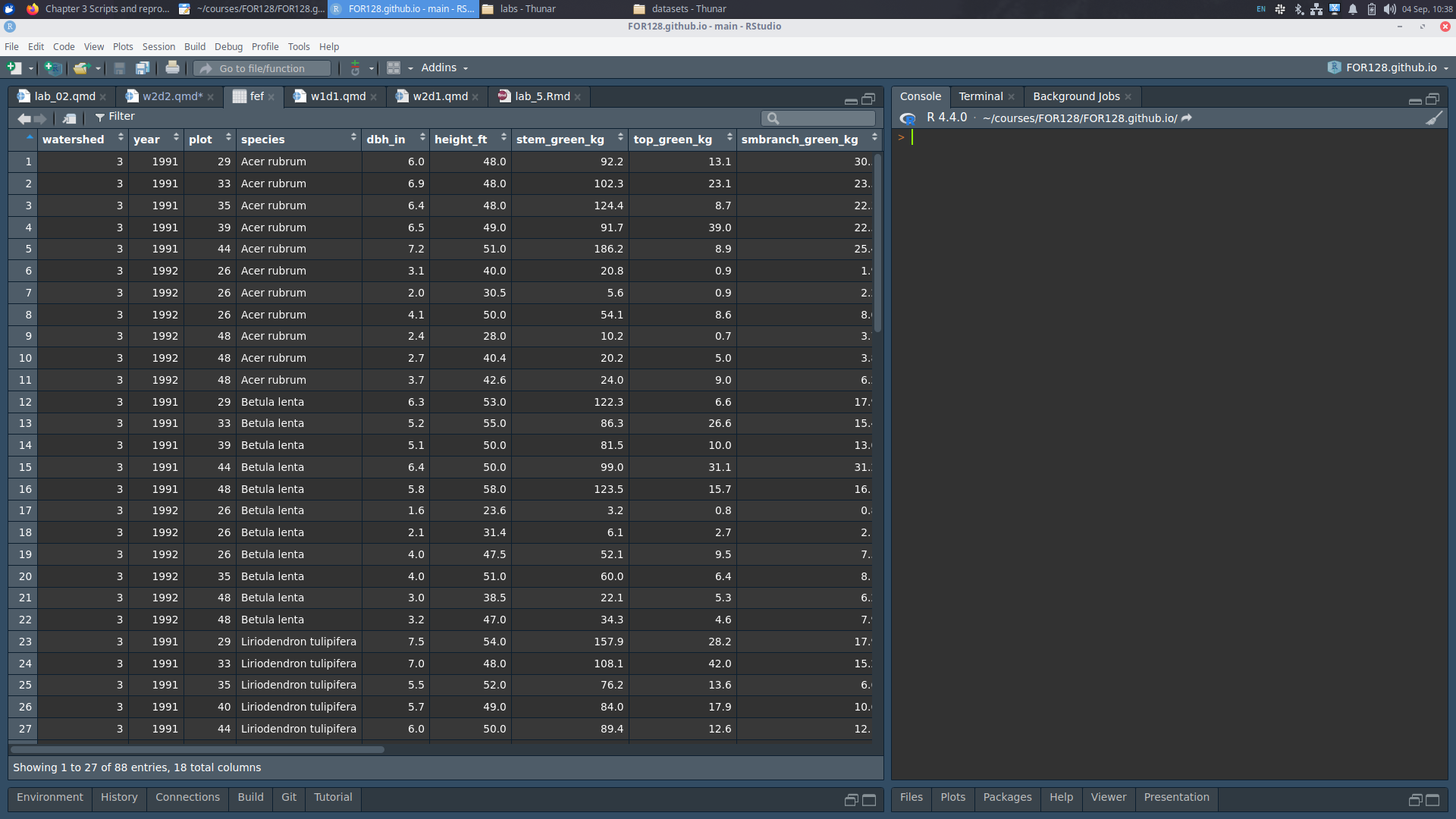
We can take a look at our dataset in R:
[1] 88 18 watershed year plot species dbh_in height_ft stem_green_kg top_green_kg
1 3 1991 29 Acer rubrum 6.0 48 92.2 13.1
2 3 1991 33 Acer rubrum 6.9 48 102.3 23.1
3 3 1991 35 Acer rubrum 6.4 48 124.4 8.7
4 3 1991 39 Acer rubrum 6.5 49 91.7 39.0
5 3 1991 44 Acer rubrum 7.2 51 186.2 8.9
6 3 1992 26 Acer rubrum 3.1 40 20.8 0.9
smbranch_green_kg lgbranch_green_kg allwoody_green_kg leaves_green_kg
1 30.5 48.4 184.2 16.1
2 23.5 57.7 206.6 12.9
3 22.3 44.1 199.5 16.5
4 22.5 35.5 188.7 12.0
5 25.4 65.1 285.6 22.4
6 1.9 1.5 25.1 0.9
stem_dry_kg top_dry_kg smbranch_dry_kg lgbranch_dry_kg allwoody_dry_kg
1 54.7 7.1 15.3 28.0 105.1
2 62.3 12.4 14.8 33.6 123.1
3 73.3 4.6 11.5 25.1 114.4
4 53.6 21.3 11.2 19.8 105.9
5 106.4 4.7 11.7 36.1 159.0
6 11.7 0.5 1.1 0.9 14.2
leaves_dry_kg
1 6.1
2 4.6
3 6.1
4 4.2
5 7.9
6 0.3Reading data
read.csv() reads comma separated value (csv) files, but R has the ability to read a massive variety of file types:
read.table()allows you to read file types with different separators like tabs and spaces, rather than just commas.
Reading data
If we wanted to use read.table() to load in FEF_trees.csv, we would run the following code:
Reading data
If we wanted to use read.table() to load in FEF_trees.csv, we would run the following code:
Reading data
If we wanted to use read.table() to load in FEF_trees.csv, we would run the following code:
Reading data: tab separated
We also have the file FEF_trees.tsv, which looks a bit different than FEF_trees.csv.
FEF_trees.csv

FEF_trees.tsv
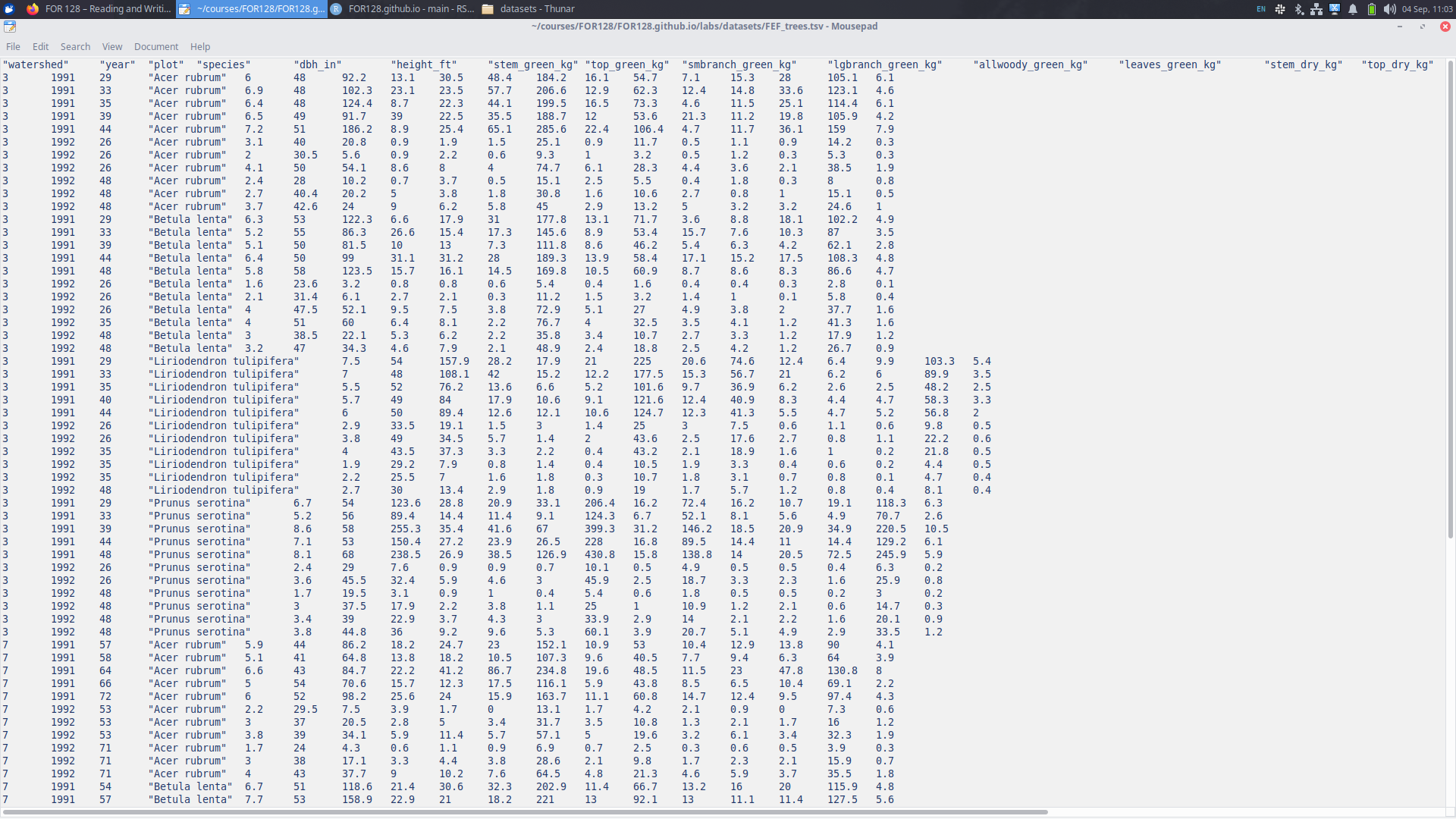
Reading FEF_trees.tsv into R
Reading FEF_trees.tsv into R
Writing data
Similar to
read.csv()andread.table(),Rincludeswrite.csv()andwrite.table()functions.These functions allow you to create (or “write”) your own files with an R object.
Let’s take a look!
Writing data
First, we look at the files in my “labs” folder
[1] "data_lab9" "data_lab9.zip" "datasets"
[4] "FEF_trees.csv" "lab_01_solutions.pdf" "lab_01_solutions.qmd"
[7] "lab_01.qmd" "lab_02.qmd" "lab_03_example.R"
[10] "lab_03.html" "lab_03.qmd" "lab_04.qmd"
[13] "lab_05.qmd" "lab_06.qmd" "lab_07_web.html"
[16] "lab_07_web.qmd" "lab_07.qmd" "lab_08_web.html"
[19] "lab_08_web.qmd" "lab_08.qmd" "lab_09_web.html"
[22] "lab_09_web.qmd" "lab_09.qmd" "lab_10_web_files"
[25] "lab_10_web.html" "lab_10_web.qmd" "lab_10.qmd"
[28] "lab_11.qmd" Writing data
Next, we can write the fef object to a .csv file.
Writing data
Next, we can write the fef object to a .csv file.
Now, we can see that the file is written.
[1] "data_lab9" "data_lab9.zip" "datasets"
[4] "FEF_trees.csv" "lab_01_solutions.pdf" "lab_01_solutions.qmd"
[7] "lab_01.qmd" "lab_02.qmd" "lab_03_example.R"
[10] "lab_03.html" "lab_03.qmd" "lab_04.qmd"
[13] "lab_05.qmd" "lab_06.qmd" "lab_07_web.html"
[16] "lab_07_web.qmd" "lab_07.qmd" "lab_08_web.html"
[19] "lab_08_web.qmd" "lab_08.qmd" "lab_09_web.html"
[22] "lab_09_web.qmd" "lab_09.qmd" "lab_10_web_files"
[25] "lab_10_web.html" "lab_10_web.qmd" "lab_10.qmd"
[28] "lab_11.qmd" "my_fef.csv" Next Week
- Andy will deliver lecture and lab through Tuesday 9/17.
- Main focuses: Chapter 4 (Data Structures).